8F1U
 
 | | Structure of a 24mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (13.8 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F0A
 
 | | Client-bound structure of a DegP trimer within a 12mer cage | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
6VGQ
 
 | | ClpP1P2 complex from M. tuberculosis with GLF-CMK bound to ClpP1 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, Z-Gly-leu-phe-CH2Cl | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VGK
 
 | | ClpP1P2 complex from M. tuberculosis | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VFX
 
 | | ClpXP from Neisseria meningitidis - Conformation B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
6VFS
 
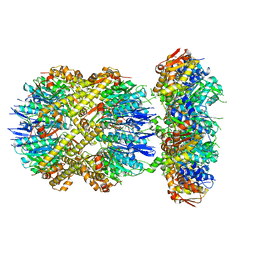 | | ClpXP from Neisseria meningitidis - Conformation A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
6VGN
 
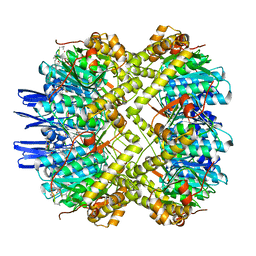 | | ClpP1P2 complex from M. tuberculosis bound to ADEP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, R0M-WFP-ALO-PRO-YCP-ALA-MP8 | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DKF
 
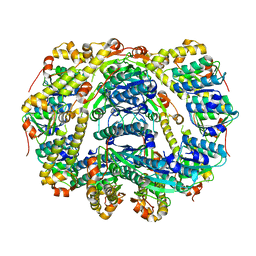 | |
1THQ
 
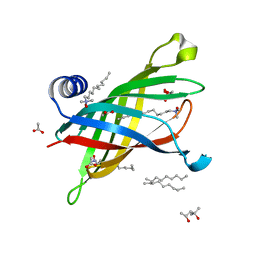 | | Crystal Structure of Outer Membrane Enzyme PagP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CrcA protein, ... | | Authors: | Ahn, V.E, Lo, E.I, Engel, C.K, Chen, L, Hwang, P.M, Kay, L.E, Bishop, R.E, Prive, G.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin.
Embo J., 23, 2004
|
|
1TBA
 
 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | Authors: | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
2K3B
 
 | |
2JSS
 
 | | NMR structure of chaperone Chz1 complexed with histone H2A.Z-H2B | | Descriptor: | Chimera of Histone H2B.1 and Histone H2A.Z, Uncharacterized protein YER030W | | Authors: | Zhou, Z, Feng, H, Hansen, D.F, Kato, H, Luk, E, Freedberg, D.I, Kay, L.E, Wu, C, Bai, Y. | | Deposit date: | 2007-07-11 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2KU2
 
 | |
2LKS
 
 | | Ff11-60 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Barette, J, Velyvis, A, Religa, T.L, Korzhnev, D.M, Kay, L.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cross-Validation of the Structure of a Transiently Formed and Low Populated FF Domain Folding Intermediate Determined by Relaxation Dispersion NMR and CS-Rosetta.
J.Phys.Chem.B, 116, 2012
|
|
2KU1
 
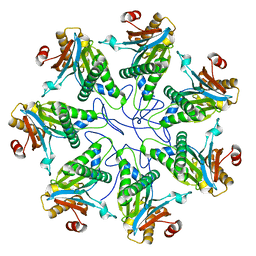 | |
2L9V
 
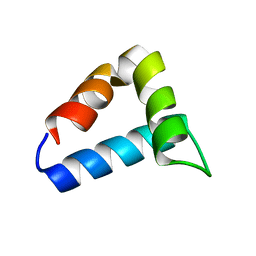 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
1ETG
 
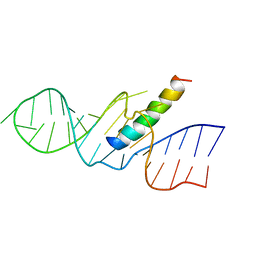 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, 19 STRUCTURES | | Descriptor: | REV PEPTIDE, REV RESPONSIVE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1ETF
 
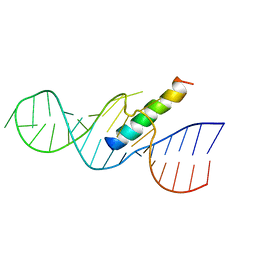 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REV PEPTIDE, REV RESPONSE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1EXH
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DZ5
 
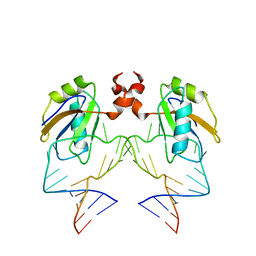 | | The NMR structure of the 38KDa U1A protein-PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein | | Descriptor: | PIE, RNA (5'-R(*GP*AP*GP*AP*CP*AP*UP*UP*GP*CP*AP*CP*CP* CP*GP*GP*AP*GP*UP*CP*UP*C)-3'), U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Varani, L, Gunderson, S.I, Mattaj, I.W, Kay, L.E, Neuhaus, D, Varani, G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-03-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the 38kDa U1A Protein-Pie RNA Complex Reveals the Basis of Cooperativity in Regulation of Polyadenylation by Human U1A Protein
Nat.Struct.Biol., 7, 2000
|
|
1EXG
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
2LCB
 
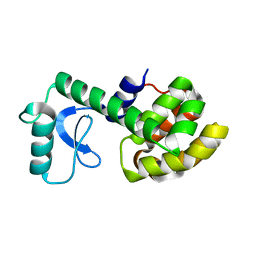 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
2L2P
 
 | | Folding Intermediate of the Fyn SH3 A39V/N53P/V55L from NMR Relaxation Dispersion Experiments | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2010-08-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
2LP5
 
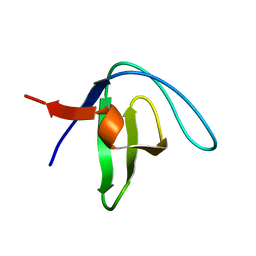 | | Native Structure of the Fyn SH3 A39V/N53P/V55L | | Descriptor: | Tyrosine-protein kinase Fyn | | Authors: | Neudecker, P, Robustelli, P, Cavalli, A, Vendruscolo, M, Kay, L.E. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an intermediate state in protein folding and aggregation.
Science, 336, 2012
|
|
2LC9
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
