8PJB
 
 | |
8PJJ
 
 | |
6Q6E
 
 | | Structural and functional insights into the condensin ATPase cycle | | Descriptor: | Condensin complex subunit 2,Structural maintenance of chromosomes protein,Structural maintenance of chromosomes protein | | Authors: | Simon, B, Hassler, M, Haering, C.H, Hennig, J. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
7BJS
 
 | | Crystal structure of Khc/atypical Tm1 complex | | Descriptor: | Kinesin heavy chain, SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJN
 
 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 270-334 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7BJG
 
 | | Crystal structure of atypical Tm1 (Tm1-I/C), residues 262-363 | | Descriptor: | SD21996p | | Authors: | Dimitrova-Paternoga, L, Jagtap, P.K.A, Ephrussi, A, Hennig, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis of mRNA transport by a kinesin-1-atypical tropomyosin complex.
Genes Dev., 35, 2021
|
|
7O0B
 
 | |
6I3R
 
 | | Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1/2 of Drosophila helicase MLE | | Descriptor: | Dosage compensation regulator | | Authors: | Jagtap, P.K.A, Buelow, S.v, Masiewicz, P, Simon, B, Hennig, J. | | Deposit date: | 2018-11-07 | | Release date: | 2019-02-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless.
Nucleic Acids Res., 47, 2019
|
|
6Y96
 
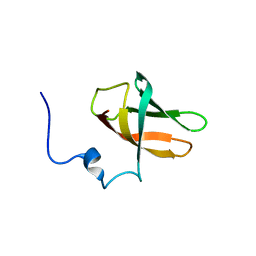 | | solution structure of cold-shock domain 9 of drosophila Upstream of N-Ras (Unr) | | Descriptor: | Upstream of N-ras, isoform A | | Authors: | Sweetapple, L.J, Hollmann, N.M, Simon, B, Hennig, J. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep, 32, 2020
|
|
6Y6E
 
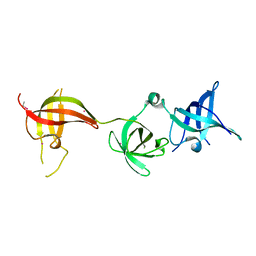 | | drosophila Unr CSD456 | | Descriptor: | ETHYL MERCURY ION, Upstream of N-ras, isoform A | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep, 32, 2020
|
|
6Y6M
 
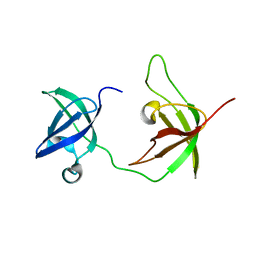 | |
6Y4H
 
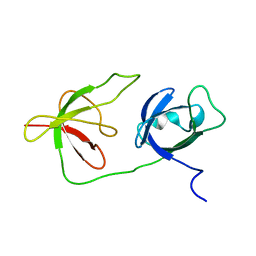 | |
4GZI
 
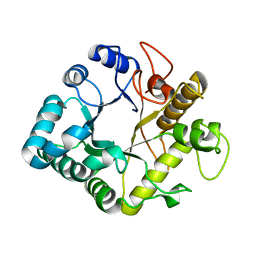 | | Active-site mutant of potato endo-1,3-beta-glucanase in complex with laminaratriose | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2012-09-06 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structures of an active-site mutant of a plant 1,3-beta-glucanase in complex with oligosaccharide products of hydrolysis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GZJ
 
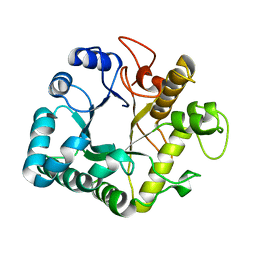 | | Active-site mutant of potato endo-1,3-beta-glucanase in complex with laminaratriose and laminaratetrose | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2012-09-06 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of an active-site mutant of a plant 1,3-beta-glucanase in complex with oligosaccharide products of hydrolysis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6FWN
 
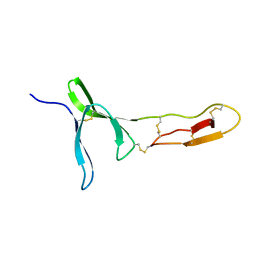 | | Structure and dynamics of the platelet integrin-binding C4 domain of von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Xu, E.-R, von Buelow, S, Chen, P.-C, Lenting, P.J, Kolsek, K, Aponte-Santamaria, C, Simon, B, Foot, J, Obser, T, Graeter, F, Schneppenheim, R, Denis, C.V, Wilmanns, M, Hennig, J. | | Deposit date: | 2018-03-06 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the platelet integrin-binding C4 domain of von Willebrand factor.
Blood, 133, 2019
|
|
5NV8
 
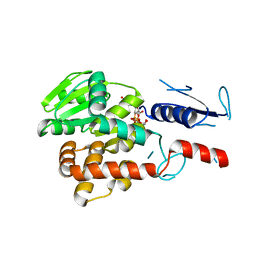 | | Structural basis for EarP-mediated arginine glycosylation of translation elongation factor EF-P | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, EF-P arginine 32 rhamnosyl-transferase | | Authors: | Macosek, J, Krafczyk, R, Jagtap, P.K.A, Lassaka, J, Hennig, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural Basis for EarP-Mediated Arginine Glycosylation of Translation Elongation Factor EF-P.
MBio, 8, 2017
|
|
5O3J
 
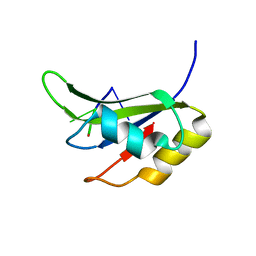 | | Crystal structure of TIA-1 RRM2 in complex with RNA | | Descriptor: | Nucleolysin TIA-1 isoform p40, RNA (5'-R(P*UP*UP*C)-3') | | Authors: | Sonntag, M, Jagtap, P.K.A, Hennig, J, Sattler, M. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7OCZ
 
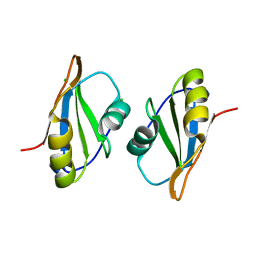 | |
7OCX
 
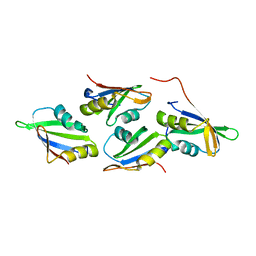 | |
7O6N
 
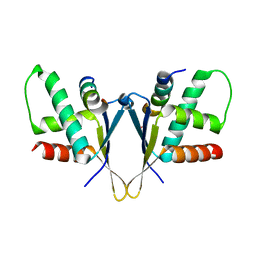 | | Crystal structure of C. elegans ERH-2 PID-3 complex | | Descriptor: | Enhancer of rudimentary homolog 2, FORMIC ACID, Protein pid-3 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7O6L
 
 | | Crystal structure of C. elegans ERH-2 | | Descriptor: | Enhancer of rudimentary homolog 2 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
6QJ3
 
 | | Crystal structure of the C. thermophilum condensin Ycs4-Brn1 subcomplex | | Descriptor: | Brn1, Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2 | | Authors: | Hassler, M, Haering, C.H, Kschonsak, M. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
6QJ1
 
 | |
4ZLR
 
 | | Structure of the Brat-NHL domain bound to consensus RNA motif | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Brain tumor protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Jakob, L, Treiber, N, Treiber, T, Stotz, M, Loedige, I, Meister, G. | | Deposit date: | 2015-05-01 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain-Tumor-Mediated Gene Regulation.
Cell Rep, 13, 2015
|
|
5E4X
 
 | | Crystal structure of cpSRP43 chromodomain 3 | | Descriptor: | MAGNESIUM ION, Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
