8K9X
 
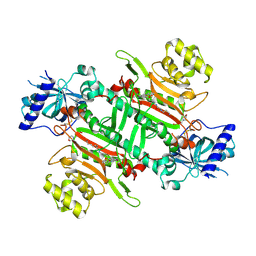 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 5 (ADKI5) | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-[3-[2-[[4-[(2,5-dimethoxyphenyl)amino]-1,3,5-triazin-2-yl]amino]phenyl]sulfonylpropyl]hexanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lysine--tRNA ligase | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9V
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 3 (ADKI3) | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9S
 
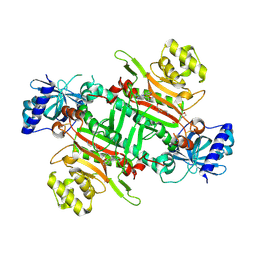 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 1 (ADKI1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9W
 
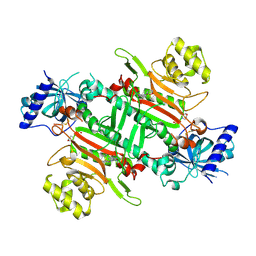 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 4 (ADKI4) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lysine--tRNA ligase, ~{N}2-(2-methoxyphenyl)-~{N}4-(2-propan-2-ylsulfonylphenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9U
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 2 (ADKI2) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
7YDG
 
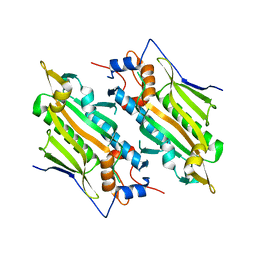 | | Crystal structure of human SARS2 catalytic domain with a disease related mutation | | Descriptor: | Serine--tRNA ligase, mitochondrial | | Authors: | Wu, S, Li, P, Zhou, X.L, Fang, P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
7YDF
 
 | | Crystal structure of human SARS2 catalytic domain | | Descriptor: | Serine--tRNA ligase, mitochondrial | | Authors: | Wu, S, Li, P, Zhou, X.L, Fang, P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
5ED6
 
 | | crystal structure of human Hint1 H114A mutant complexing with ATP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2015-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
8H99
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8H9C
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 4 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8H98
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 1 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8H9B
 
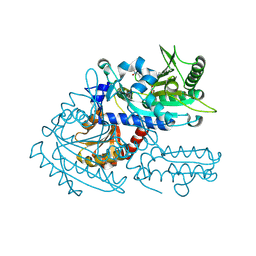 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 3 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8H9A
 
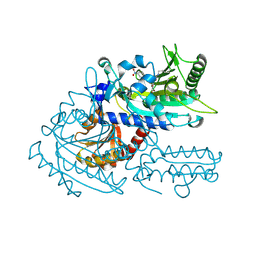 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 2 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
5ED3
 
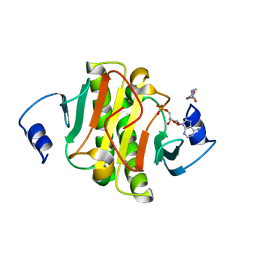 | | crystal structure of human Hint1 complexing with AP5A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2015-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
8HK0
 
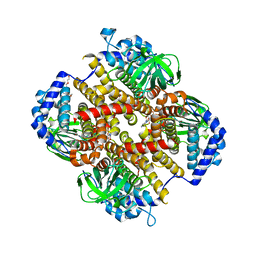 | | Crystal structure of Fic32-33 complex from Streptomyces ficellus NRRL 8067 | | Descriptor: | Acyl-CoA dehydrogenase, Dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cheng, Y, Qiao, H, Liu, W, Fang, P. | | Deposit date: | 2022-11-24 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
J.Am.Chem.Soc., 145, 2023
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | Descriptor: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | Authors: | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
4DKT
 
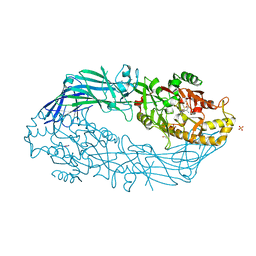 | | Crystal structure of human peptidylarginine deiminase 4 in complex with N-acetyl-L-threonyl-L-alpha-aspartyl-N5-[(1E)-2-fluoroethanimidoyl]-L-ornithinamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Jones, J.E, Slack, J.L, Fang, P, Zhang, X, Subramanian, V, Causey, C.P, Coonrod, S.A, Guo, M, Thompson, P.R. | | Deposit date: | 2012-02-04 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Synthesis and Screening of a Haloacetamidine Containing Library To Identify PAD4 Selective Inhibitors.
Acs Chem.Biol., 7, 2012
|
|
4N22
 
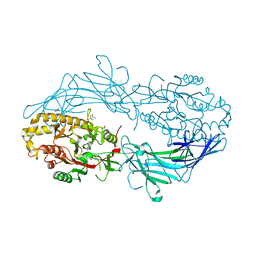 | | Crystal structure of Protein Arginine Deiminase 2 (50 uM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2I
 
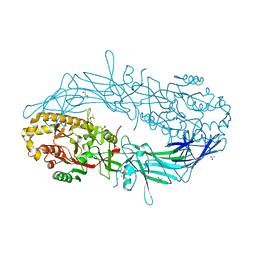 | | Crystal structure of Protein Arginine Deiminase 2 (D177A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N26
 
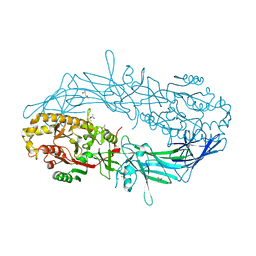 | | Crystal structure of Protein Arginine Deiminase 2 (500 uM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2L
 
 | | Crystal structure of Protein Arginine Deiminase 2 (Q350A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2D
 
 | | Crystal structure of Protein Arginine Deiminase 2 (D123N, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2B
 
 | | Crystal structure of Protein Arginine Deiminase 2 (10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2A
 
 | | Crystal structure of Protein Arginine Deiminase 2 (5 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
