2XSK
 
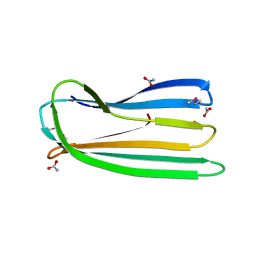 | | E. coli curli protein CsgC - SeCys | | Descriptor: | ACETATE ION, CSGC | | Authors: | Salgado, P.S, Taylor, J.D, Cota, E, Matthews, S.J. | | Deposit date: | 2010-09-29 | | Release date: | 2010-12-29 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the Usability of the Phasing Power of Diselenide Bonds: Secys Sad Phasing of Csgc Using a Non-Auxotrophic Strain.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2WNM
 
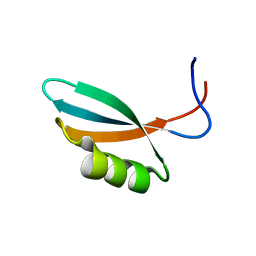 | | Solution structure of Gp2 | | Descriptor: | GENE 2 | | Authors: | Camara, B, Liu, M, Shadrinc, A, Liu, B, Simpson, P, Weinzierl, R, Severinovc, K, Cota, E, Matthews, S, Wigneshweraraj, S.R. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | T7 Phage Protein Gp2 Inhibits the Escherichia Coli RNA Polymerase by Antagonizing Stable DNA Strand Separation Near the Transcription Start Site.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2Y7N
 
 | |
2Y7O
 
 | |
2Y7M
 
 | |
2Y7L
 
 | |
2YLH
 
 | |
6EHJ
 
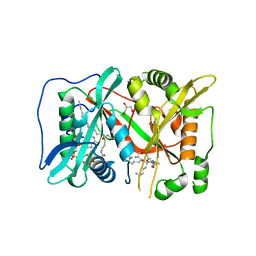 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and peptide bound | | Descriptor: | ASPARAGINE, COENZYME A, GLYCEROL, ... | | Authors: | Perez-Dorado, I, Ritzefeld, M, Tate, E.W. | | Deposit date: | 2017-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
5OJP
 
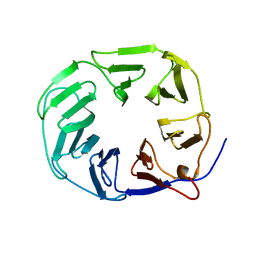 | | YCF48 bound to D1 peptide | | Descriptor: | Ycf48-like protein | | Authors: | Michoux, F, Nixon, P.J, Murray, J.W. | | Deposit date: | 2017-07-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6QRM
 
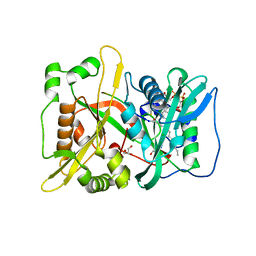 | | HsNMT1 in complex with both MyrCoA and GNCFSKRRAA substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
2LMC
 
 | |
2KUB
 
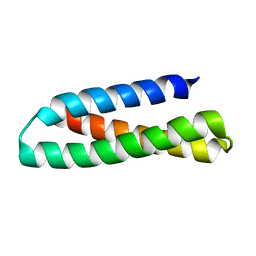 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
2X12
 
 | |
2QST
 
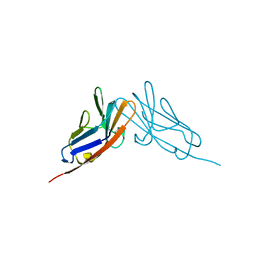 | |
5OJ3
 
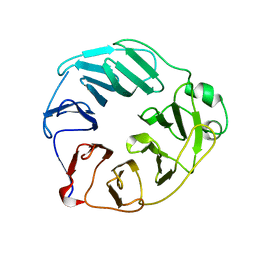 | | YCF48 from Cyanidioschyzon merolae | | Descriptor: | Photosystem II stability/assembly factor HCF136 | | Authors: | Michoux, F, Murray, J.W, Nixon, P.J. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.982 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OJR
 
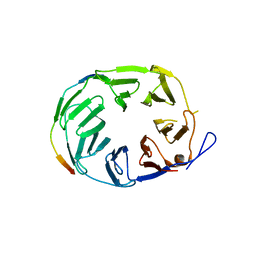 | | YCF48 bound to D1 peptide | | Descriptor: | Photosystem II protein D1 3, Ycf48-like protein | | Authors: | Michoux, F, Nixon, P.J, Murray, J.W, Bialek, W, Thieulin-Pardo, G. | | Deposit date: | 2017-07-23 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OJ5
 
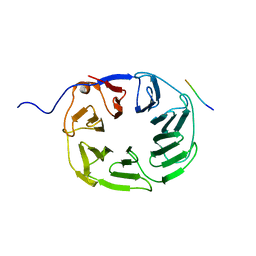 | | YCF48 bound to D1 peptide | | Descriptor: | PHE-PRO-LEU-ASP-LEU-ALA, Ycf48-like protein | | Authors: | Michoux, F, Nixon, P.J, Murray, J.W. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
2QSQ
 
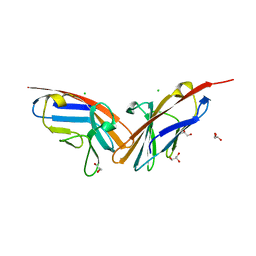 | | Crystal structure of the N-terminal domain of carcinoembryonic antigen (CEA) | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 5, GLYCEROL | | Authors: | Le Trong, I, Korotkova, N, Moseley, S.L, Stenkamp, R.E. | | Deposit date: | 2007-07-31 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of Dr adhesins of Escherichia coli to carcinoembryonic antigen triggers receptor dissociation.
Mol.Microbiol., 67, 2008
|
|
3QS2
 
 | |
3QS3
 
 | |
3SI5
 
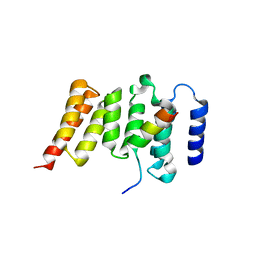 | | Kinetochore-BUBR1 kinase complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Protein CASC5 | | Authors: | Blundell, T.L, Chirgadze, D.Y, Bolanos-Garcia, V.M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Blinkin-BUBR1 Complex Reveals an Interaction Crucial for Kinetochore-Mitotic Checkpoint Regulation via an Unanticipated Binding Site.
Structure, 19, 2011
|
|
3UIY
 
 | |
3UIZ
 
 | |
6SK2
 
 | | HsNMT1 in complex with both MyrCoA and Acetylated-GKSFSKPR peptide reveals N-terminal Lysine Myristoylation | | Descriptor: | Apoptosis-inducing factor 3, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.90000653 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
6SKJ
 
 | | DeltaC2 C-terminal truncation of HsNMT1 in complex with MyrCoA and GNCFSKPR substrates | | Descriptor: | Apoptosis-inducing factor 3, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
