1OU8
 
 | | structure of an AAA+ protease delivery protein in complex with a peptide degradation tag | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog, synthetic ssrA peptide | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
4YJX
 
 | | The structure of Agrobacterium tumefaciens ClpS2 bound to L-phenylalaninamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, PHENYLALANINE AMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7M1L
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, PHOSPHATE ION | | Authors: | Hall, B.M, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
4YJM
 
 | |
4YKA
 
 | | The structure of Agrobacterium tumefaciens ClpS2 in complex with L-tyrosinamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, L-TYROSINAMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
4I5O
 
 | | Crystal Structure of W-W-R ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.4787 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I9K
 
 | | Crystal structure of symmetric W-W-W ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-12-05 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.0003 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I63
 
 | | Crystal Structure of E-R ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.709 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
1OU9
 
 | | Structure of SspB, a AAA+ protease delivery protein | | Descriptor: | CALCIUM ION, Stringent starvation protein B homolog | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1OUL
 
 | | Structure of the AAA+ protease delivery protein SspB | | Descriptor: | Stringent starvation protein B homolog | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
3DNJ
 
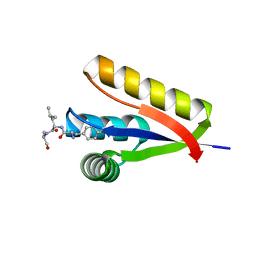 | | The structure of the Caulobacter crescentus ClpS protease adaptor protein in complex with a N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, synthetic N-end rule peptide | | Authors: | Wang, K, Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2008-07-02 | | Release date: | 2008-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The molecular basis of N-end rule recognition.
Mol.Cell, 32, 2008
|
|
4I4L
 
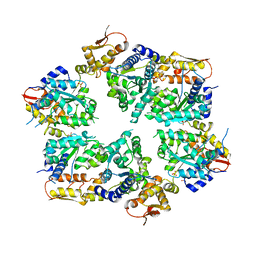 | | Crystal Structure of Nucleotide-Bound W-W-W ClpX Hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6981 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I81
 
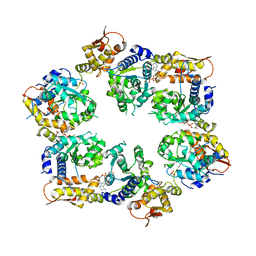 | | Crystal Structure of ATPgS bound ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-12-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8182 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
2QAZ
 
 | |
3EQ2
 
 | |
2QAS
 
 | |
4I34
 
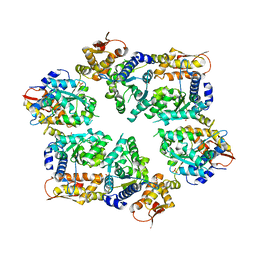 | | Crystal Structure of W-W-W ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1218 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
3ES2
 
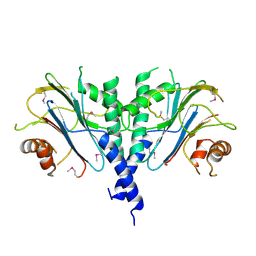 | |
3EOD
 
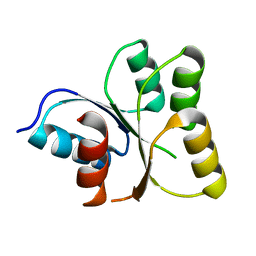 | |
5JI3
 
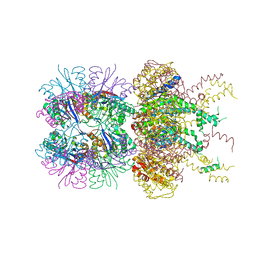 | | HslUV complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
6PP7
 
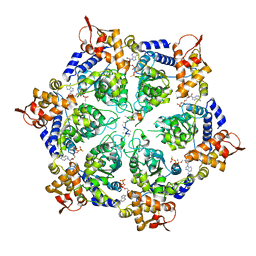 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6POD
 
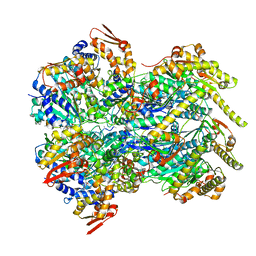 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP5
 
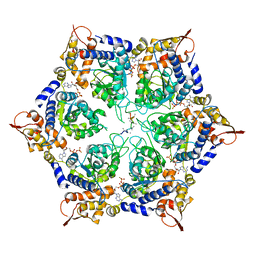 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PO3
 
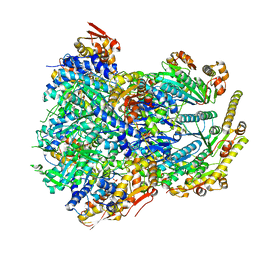 | | ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
