3CYJ
 
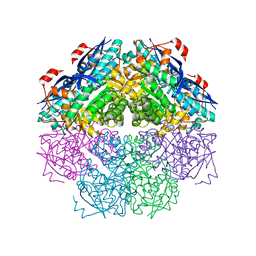 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
3CKW
 
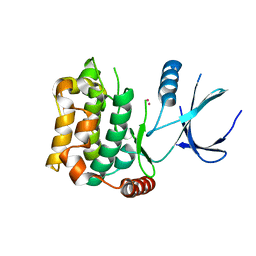 | | Crystal structure of sterile 20-like kinase 3 (MST3, STK24) | | Descriptor: | MERCURY (II) ION, Serine/threonine-protein kinase 24 | | Authors: | Antonysamy, S.S, Burley, S.K, Buchanan, S, Chau, F, Feil, I, Wu, L, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-17 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of sterile 20-like kinase 3 (MST3, STK24).
To be Published
|
|
3CTA
 
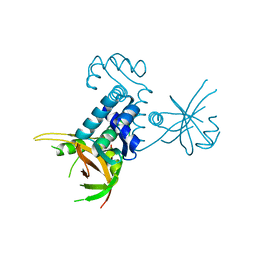 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
3CTP
 
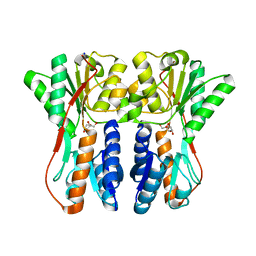 | | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with D-xylulofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, SODIUM ION, beta-D-xylulofuranose | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with L-xylulose.
To be Published
|
|
3DBI
 
 | | CRYSTAL STRUCTURE OF SUGAR-BINDING TRANSCRIPTIONAL REGULATOR (LACI FAMILY) FROM ESCHERICHIA COLI COMPLEXED WITH PHOSPHATE | | Descriptor: | GLYCEROL, PHOSPHATE ION, SUGAR-BINDING TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Wu, B, Maletic, M, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Sugar-Binding Transcriptional Regulator (LacI Family) from Escherichia Coli Complexed with Phosphate.
To be Published
|
|
3DBY
 
 | | Crystal structure of uncharacterized protein from Bacillus cereus G9241 (CSAP Target) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, uncharacterized protein | | Authors: | Ramagopal, U.A, Bonanno, J.B, Ozyurt, S, Freeman, J, Wasserman, S, Hu, S, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uncharacterized protein from Bacillus cereus G9241
To be published
|
|
3DEC
 
 | | Crystal structure of a glycosyl hydrolases family 2 protein from Bacteroides thetaiotaomicron | | Descriptor: | Beta-galactosidase, POTASSIUM ION | | Authors: | Kumaran, D, Bonanno, J, Romero, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-09 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Glycosyl Hydrolases Family 2 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
3DHU
 
 | | Crystal structure of an alpha-amylase from Lactobacillus plantarum | | Descriptor: | Alpha-amylase | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an alpha-amylase from Lactobacillus plantarum
To be Published
|
|
1SEF
 
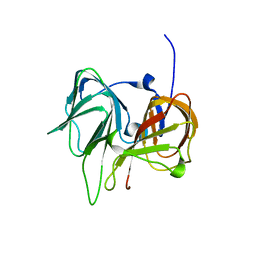 | |
1SFN
 
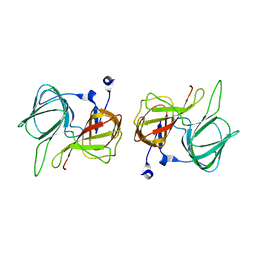 | |
1S80
 
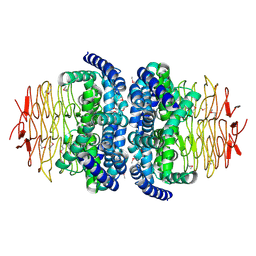 | | Structure of Serine Acetyltransferase from Haemophilis influenzae Rd | | Descriptor: | Serine acetyltransferase | | Authors: | Gorman, J, Gogos, A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-01-30 | | Release date: | 2004-08-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of serine acetyltransferase from Haemophilus influenzae Rd.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RZN
 
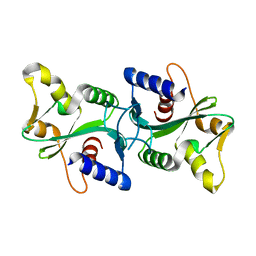 | |
1SGJ
 
 | |
1SG9
 
 | | Crystal structure of Thermotoga maritima protein HEMK, an N5-glutamine methyltransferase | | Descriptor: | GLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel mode of dimerization via formation of a glutamate anhydride crosslink in a protein crystal structure.
Proteins, 71, 2008
|
|
1SGM
 
 | |
3LVS
 
 | | Crystal structure of farnesyl diphosphate synthase from rhodobacter capsulatus sb1003 | | Descriptor: | FARNESYL DIPHOSPHATE SYNTHASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3MGK
 
 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | Descriptor: | Intracellular protease/amidase related enzyme (ThiJ family) | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
3MMZ
 
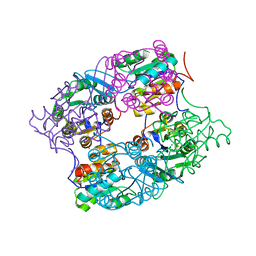 | | CRYSTAL STRUCTURE OF putative HAD family hydrolase from Streptomyces avermitilis MA-4680 | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative HAD family hydrolase | | Authors: | Malashkevich, V.N, Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3ME5
 
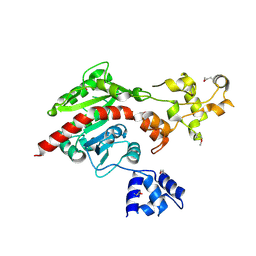 | | Crystal structure of putative dna cytosine methylase from shigella flexneri 2a str. 2457T | | Descriptor: | Cytosine-specific methyltransferase | | Authors: | Ramagopal, U.A, Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of putative dna cytosine methylase from shigella flexneri 2a str. 2457T
To be Published
|
|
3MAE
 
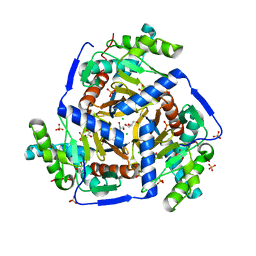 | | CRYSTAL STRUCTURE OF PROBABLE DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM LISTERIA MONOCYTOGENES 4b F2365 | | Descriptor: | 2-oxoisovalerate dehydrogenase E2 component, dihydrolipoamide acetyltransferase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A CATALYTIC DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM LISTERIA MONOCYTOGENES 4b F2365
To be Published
|
|
3LYB
 
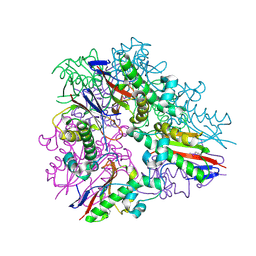 | |
3LYP
 
 | |
3ME7
 
 | | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
3ME8
 
 | | Crystal structure of putative electron transfer protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transfer protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
