5ZME
 
 | |
5ZMF
 
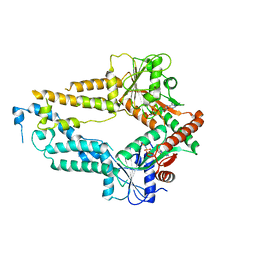 | | AMPPNP complex of C. reinhardtii ArsA1 | | Descriptor: | ATPase ARSA1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lin, T.W, Hsiao, C.D, Chang, H.Y. | | Deposit date: | 2018-04-03 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.556 Å) | | Cite: | Structural analysis of chloroplast tail-anchored membrane protein recognition by ArsA1.
Plant J., 99, 2019
|
|
1GGG
 
 | |
1AD3
 
 | |
2PGI
 
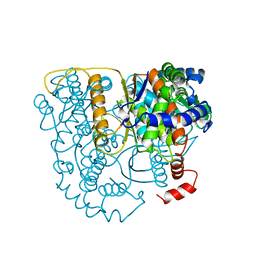 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE-AN ENZYME WITH AUTOCRINE MOTILITY FACTOR ACTIVITY IN TUMOR CELLS | | Descriptor: | PHOSPHOGLUCOSE ISOMERASE | | Authors: | Sun, Y.-J, Chou, C.-C, Chen, W.-S, Meng, M, Hsiao, C.-D. | | Deposit date: | 1998-10-27 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of a multifunctional protein: phosphoglucose isomerase/autocrine motility factor/neuroleukin.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1GSU
 
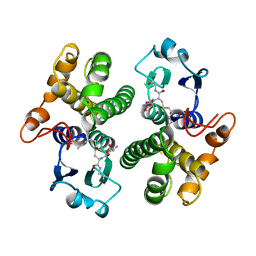 | | AN AVIAN CLASS-MU GLUTATHIONE S-TRANSFERASE, CGSTM1-1 AT 1.94 ANGSTROM RESOLUTION | | Descriptor: | CLASS-MU GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Sun, Y.-J, Kuan, C, Tam, M.F, Hsiao, C.-D. | | Deposit date: | 1997-09-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution.
J.Mol.Biol., 278, 1998
|
|
1KXI
 
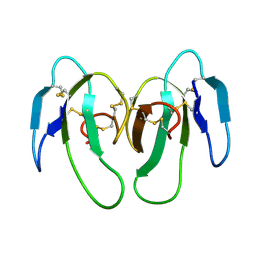 | | STRUCTURE OF CYTOTOXIN HOMOLOG PRECURSOR | | Descriptor: | CARDIOTOXIN V | | Authors: | Sun, Y.-J, Wu, W.-G, Chiang, C.-M, Hsin, A.-Y, Hsiao, C.-D. | | Deposit date: | 1996-08-29 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of cardiotoxin V from Taiwan cobra venom: pH-dependent conformational change and a novel membrane-binding motif identified in the three-finger loops of P-type cardiotoxin.
Biochemistry, 36, 1997
|
|
8JML
 
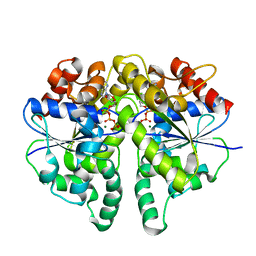 | | Structure of Helicobacter pylori Soj protein mutant, D41A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SpoOJ regulator (Soj) | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
8JMK
 
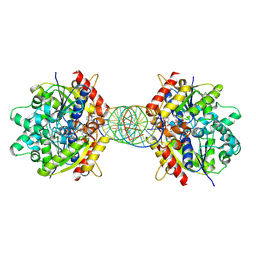 | | Structure of Helicobacter pylori Soj mutant, D41A bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
8JMJ
 
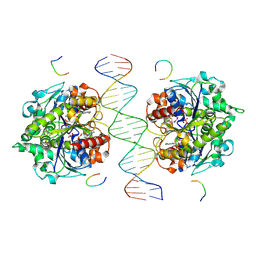 | | Structure of Helicobacter pylori Soj-DNA-Spo0J complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
1WDN
 
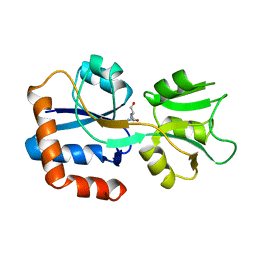 | | GLUTAMINE-BINDING PROTEIN | | Descriptor: | GLUTAMINE, GLUTAMINE BINDING PROTEIN | | Authors: | Sun, Y.-J, Rose, J, Wang, B.-C, Hsiao, C.-D. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: comparisons with other amino acid binding proteins.
J.Mol.Biol., 278, 1998
|
|
6IUB
 
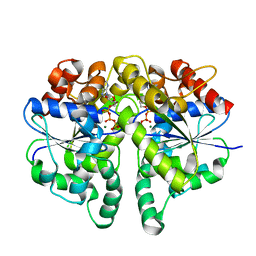 | | Structure of Helicobacter pylori Soj protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SpoOJ regulator (Soj) | | Authors: | Chu, C.H, Yen, C.Y, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
6IUD
 
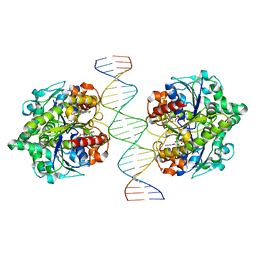 | | Structure of Helicobacter pylori Soj-ADP complex bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Yen, C.Y, Chu, C.H, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
6IUC
 
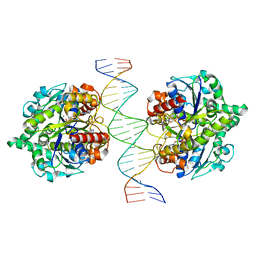 | | Structure of Helicobacter pylori Soj-ATP complex bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Chu, C.H, Yen, C.Y, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
1C7R
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | 5-PHOSPHOARABINONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
1C7Q
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | N-BROMOACETYL-AMINOETHYL PHOSPHATE, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
4UMK
 
 | | The complex of Spo0J and parS DNA in chromosomal partition system | | Descriptor: | DNA, PROBABLE CHROMOSOME-PARTITIONING PROTEIN PARB, SULFATE ION | | Authors: | Chen, B.W, Chu, C.H, Tung, J.Y, Hsu, C.E, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2014-05-19 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Insights into ParB spreading from the complex structure of Spo0J and parS.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
5EZ1
 
 | |
3K53
 
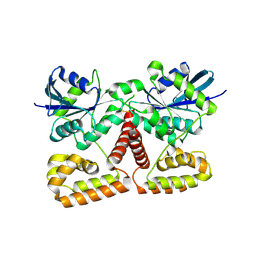 | | Crystal Structure of NFeoB from P. furiosus | | Descriptor: | Ferrous iron transport protein b | | Authors: | Eng, E.T, Dong, G, Unger, V.M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural fold, conservation and Fe(II) binding of the intracellular domain of prokaryote FeoB.
J.Struct.Biol., 170, 2010
|
|
1NPO
 
 | |
3W9S
 
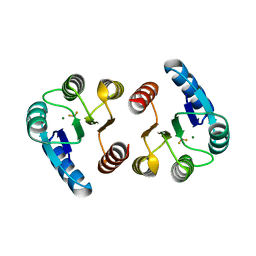 | | Crystal Structure Analysis of the N-terminal Receiver domain of Response Regulator PmrA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, OmpR family response regulator in two-component regulatory system with BasS | | Authors: | Chen, C, Luo, S. | | Deposit date: | 2013-04-15 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of a Physical Blockage Mechanism for the Interaction of Response Regulator PmrA with Connector Protein PmrD from Klebsiella Pneumoniae
J.Biol.Chem., 288, 2013
|
|
4A7W
 
 | | Crystal structure of uridylate kinase from Helicobacter pylori | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, URIDYLATE KINASE | | Authors: | Chu, C.H, Chen, P.C, Liu, M.H, Sun, Y.J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Helicobacter Pylori Uridylate Kinase: Insight Into Release of the Product Udp
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A7X
 
 | | Crystal structure of uridylate kinase from Helicobacter pylori | | Descriptor: | URIDINE-5'-DIPHOSPHATE, URIDYLATE KINASE | | Authors: | Chu, C.H, Liu, M.H, Chen, P.C, Sun, Y.J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of Helicobacter Pylori Uridylate Kinase: Insight Into Release of the Product Udp
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5ZW8
 
 | | PigA with FAD and proline | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Lee, C.-C, Ko, T.-P, Wang, A.H.J. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Crystal Structure of PigA: A Prolyl Thioester-Oxidizing Enzyme in Prodigiosin Biosynthesis.
Chembiochem, 20, 2019
|
|
5ZW2
 
 | | FAD complex of PigA | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ACETATE ION, ... | | Authors: | Lee, C.-C, Ko, T.-P, Wang, A.H.J. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal Structure of PigA: A Prolyl Thioester-Oxidizing Enzyme in Prodigiosin Biosynthesis.
Chembiochem, 20, 2019
|
|
