1DJW
 
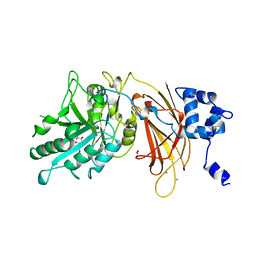 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHONATE | | Descriptor: | ACETATE ION, CALCIUM ION, INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1DJZ
 
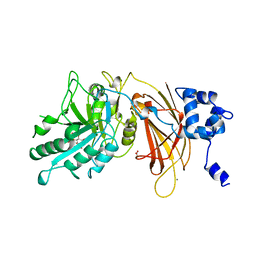 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-4,5-BISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-4,5-BISPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1DJH
 
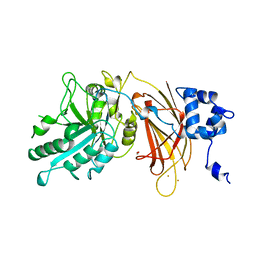 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH BARIUM | | Descriptor: | ACETATE ION, BARIUM ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
6Y98
 
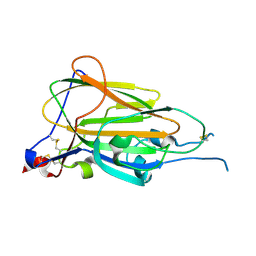 | | Crystal Structure of subtype-switched Epithelial Adhesin 9 to 1 A domain (Epa9-CBL2Epa1) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, PA14 domain-containing protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
6Y9J
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 9 A domain (Epa1-CBL2Epa9) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epa1p, ... | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
5ZM0
 
 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Gusti Ngurah Putu, E.P, Maestre-Reyna, M, Tsai, M.-D, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-03-31 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii
Nucleic Acids Res., 46, 2018
|
|
6IAM
 
 | |
6CMV
 
 | | Crystal structure of Archaeal Biofilm Regulator (AbfR2) from Sulfolobus acidocaldarius | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Transcriptional regulator Lrs14-like protein | | Authors: | Essen, L.-O, Vogt, M.S, Banerjee, A. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of an Lrs14-like archaeal biofilm regulator from Sulfolobus acidocaldarius.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7F8T
 
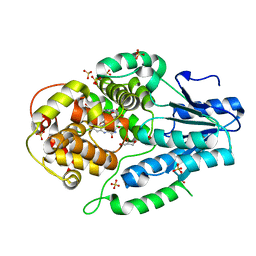 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
5ZCW
 
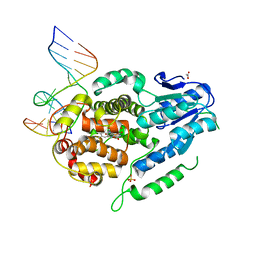 | | Structure of the Methanosarcina mazei class II CPD-photolyase in complex with intact, phosphodiester linked, CPD-lesion | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*(TTD)P*CP*GP*CP*GP*CP*AP*A)-3', 5'-D(*TP*GP*CP*GP*CP*GP*AP*AP*GP*CP*CP*GP*AP*T)-3', ACETATE ION, ... | | Authors: | Maestre-Reyna, M, Bessho, Y. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA.
IUCrJ, 5, 2018
|
|
7PZA
 
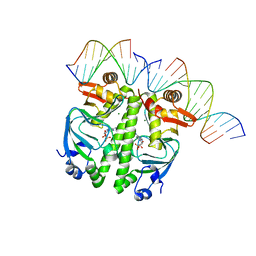 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
7PZB
 
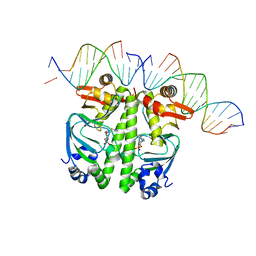 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
8PHA
 
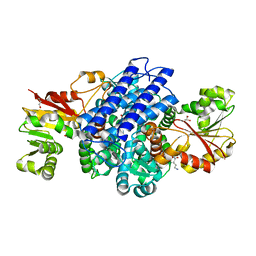 | | O(S)-methyltransferase from Pleurotus sapidus | | Descriptor: | 2-HYDROXY BUTANE-1,4-DIOL, GLYCEROL, L-ornithine, ... | | Authors: | Korf, L, Essen, L.-O. | | Deposit date: | 2023-06-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Novel O - and S -Methyltransferase from Pleurotus sapidus Is Involved in Flavor Formation.
J.Agric.Food Chem., 72, 2024
|
|
3CKY
 
 | |
6FN3
 
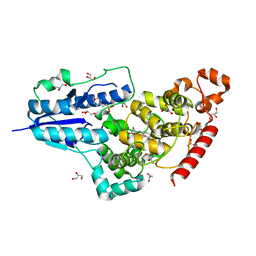 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6FN0
 
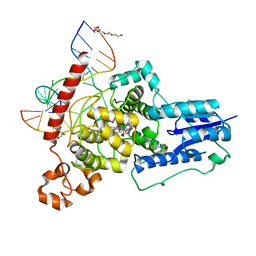 | | The animal-like Cryptochrome from Chlamydomonas reinhardtii in complex with 6-4 DNA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
5O86
 
 | |
6FN2
 
 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
5O8E
 
 | | Mutant of class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ignatz, E, Geisselbrecht, Y, Essen, L.-O. | | Deposit date: | 2017-06-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nicotinamide Adenine Dinucleotides Arrest Photoreduction of Class II DNA Photolyases in FADH ̇ State.
Photochem. Photobiol., 94, 2018
|
|
5O8D
 
 | |
5OW0
 
 | | Crystal structure of an electron transfer flavoprotein from Geobacter metallireducens | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein, alpha subunit, ... | | Authors: | Essen, L.-O, Vogt, M.S, Heider, J, Koelzer, S, Peschke, P, Chowdhury, N.P, Schuehle, K, Kleinsorge, D. | | Deposit date: | 2017-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of an electron transfer flavoprotein involved in toluene degradation in strictly anaerobic bacteria.
J.Bacteriol., 2019
|
|
6HOS
 
 | | Structure of the KpFlo2 adhesin domain in complex with glycerol | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, BA75_04148T0, CALCIUM ION, ... | | Authors: | Essen, L.-O, Kock, M, Veelders, M. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of PA14/Flo5-Like Adhesins FromKomagataella pastoris.
Front Microbiol, 9, 2018
|
|
5FV6
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | Descriptor: | ACETATE ION, Flocculation protein FLO11, GLYCEROL, ... | | Authors: | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
5FV5
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | Descriptor: | ACETATE ION, Flocculation protein FLO11 | | Authors: | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
1ASX
 
 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
