2IQY
 
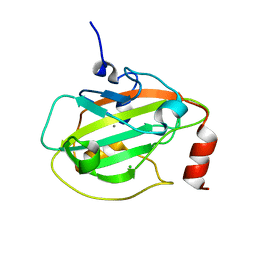 | | Rat Phosphatidylethanolamine-Binding Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phosphatidylethanolamine-binding protein 1 | | Authors: | Kim, Y, Joachimiak, G, Heil, G.L, Koide, S, Joachimiak, A. | | Deposit date: | 2006-10-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Rat Phosphatidylethanolamine-Binding Protein
To be Published
|
|
6WGP
 
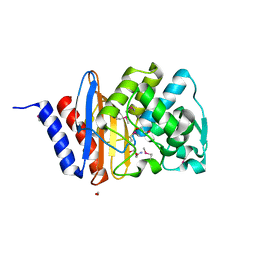 | | The crystal structure of a beta lactamase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a beta lactamase from Xanthomonas campestris pv. campestris str. ATCC 33913
To Be Published
|
|
6WRH
 
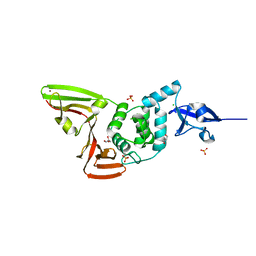 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
2IQX
 
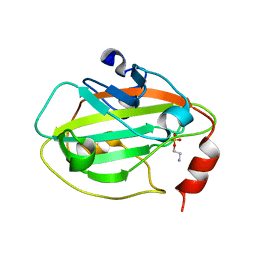 | | Rat Phosphatidylethanolamine-Binding Protein Containing the S153E Mutation in the Complex with o-Phosphorylethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Phosphatidylethanolamine-binding protein 1 | | Authors: | Kim, Y, Joachimiak, G, Clark, M.C, Rosner, M, Joachimiak, A. | | Deposit date: | 2006-10-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rat Phosphatidylethanolamine-Binding Crystal structure of Protein Containing the S153E Mutation in the Complex with o-Phosphorylethanolamine
To be Published
|
|
6X9Y
 
 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | Descriptor: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
6XFS
 
 | | Class C beta-lactamase from Escherichia coli in complex with Tazobactam | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Class C beta-lactamase from Escherichia coli in complex with Tazobactam
To Be Published
|
|
4Q6W
 
 | | Crystal Structure of Periplasmic Binding Protein type 1 from Bordetella pertussis Tohama I complexed with 3-Hydroxy Benzoic Acid | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-23 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Periplasmic Binding Protein type 1 from Bordetella pertussis Tohama I complexed with 3-Hydroxy Benzoic Acid
To be Published, 2014
|
|
4Q2B
 
 | | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-D-glucanase, FORMIC ACID, ... | | Authors: | Tan, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-25 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440
To be Published
|
|
1U14
 
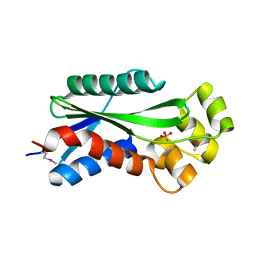 | | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom | | Descriptor: | Hypothetical UPF0244 protein yjjX, PHOSPHATE ION | | Authors: | Qiu, Y, Kim, Y, Cuff, M, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom
To be Published
|
|
4J01
 
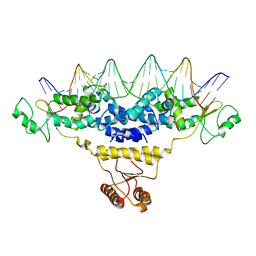 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 29mer DNA target | | Descriptor: | DNA (29-MER), SULFATE ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.246 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6XJ3
 
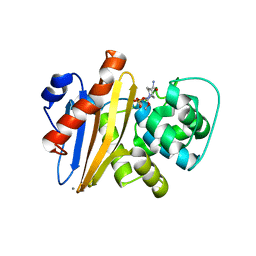 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
4J00
 
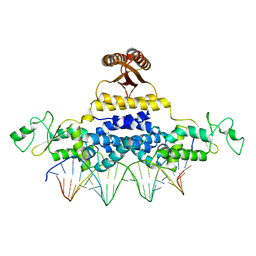 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 24mer DNA target | | Descriptor: | DNA (5'-D(*TP*GP*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*CP*C)-3'), MAGNESIUM ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1S6Y
 
 | | 2.3A crystal structure of phospho-beta-glucosidase | | Descriptor: | 6-phospho-beta-glucosidase | | Authors: | Tereshko, V, Dementieva, I, Kim, Y, Collat, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3A CRYSTAL STRUCTURE OF PHOSPHO-BETA-GLUCOSIDASE, licH Gene Product from BACILLUS STEAROTHERMOPHILUS
To be Published
|
|
6XOA
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation
To Be Published
|
|
4RGR
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, GLYCEROL, Repressor protein, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP
To be Published, 2014
|
|
4RGS
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with Vanilin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, GLYCEROL, Repressor protein, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with Vanilin
To be Published, 2014
|
|
4RGU
 
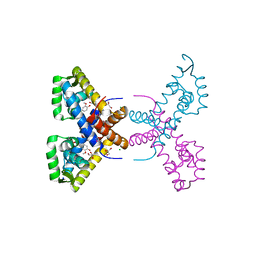 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with ferulic acid
To be Published, 2014
|
|
6BAL
 
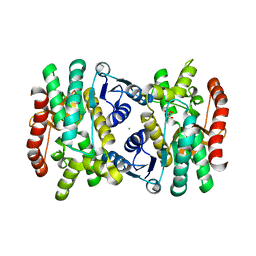 | | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CHLORIDE ION, Malate dehydrogenase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate
To Be Published
|
|
4RGX
 
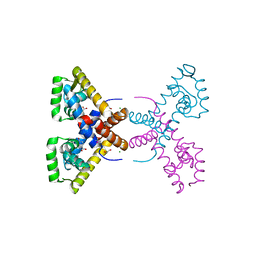 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 3,4-dihydroxy bezoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 3,4-dihydroxy bezoic acid
To be Published, 2014
|
|
6XKF
 
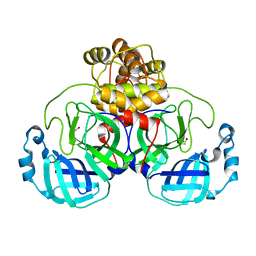 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine). | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevskyi, A.Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine).
To Be Published
|
|
6XG1
 
 | | Class C beta-lactamase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Class C beta-lactamase from Escherichia coli
To Be Published
|
|
6B4O
 
 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | Authors: | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
6B8W
 
 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | Descriptor: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
1S3X
 
 | | The crystal structure of the human Hsp70 ATPase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Heat shock 70 kDa protein 1, ... | | Authors: | Sriram, M, Osipiuk, J, Freeman, B, Morimoto, R.I, Joachimiak, A. | | Deposit date: | 2004-01-14 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain
Structure, 5, 1997
|
|
1S9U
 
 | | Atomic structure of a putative anaerobic dehydrogenase component | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, putative component of anaerobic dehydrogenases | | Authors: | Qiu, Y, Zhang, R, Tereshko, V, Kim, Y, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Proteins, 71, 2008
|
|
