5AFF
 
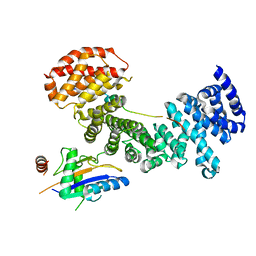 | | Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA binding site | | Descriptor: | RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L5, SYMPORTIN 1 | | Authors: | Calvino, F.R, Kharde, S, Wild, K, Bange, G, Sinning, I. | | Deposit date: | 2015-01-21 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Symportin 1 Chaperones 5S Rnp Assembly During Ribosome Biogenesis by Occupying an Essential Rrna-Binding Site.
Nat.Commun., 6, 2015
|
|
3L5D
 
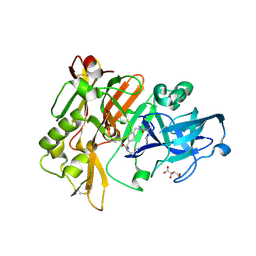 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
9HLK
 
 | | X-ray structure of the adduct formed upon reaction of the diiodido analogue of picoplatin with lysozyme (structure A) | | Descriptor: | ACETATE ION, AMMONIA, CHLORIDE ION, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2024-12-05 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Cytotoxicity and Binding to DNA, Lysozyme, Ribonuclease A, and Human Serum Albumin of the Diiodido Analog of Picoplatin.
Inorg.Chem., 64, 2025
|
|
3KMY
 
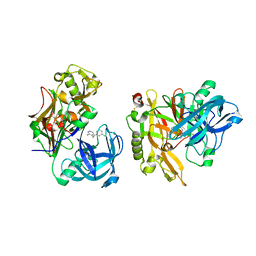 | | Structure of BACE bound to SCH12472 | | Descriptor: | 3-[2-(3-chlorophenyl)ethyl]pyridin-2-amine, Beta-secretase 1 | | Authors: | Strickland, C, Wang, Y. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
7QRA
 
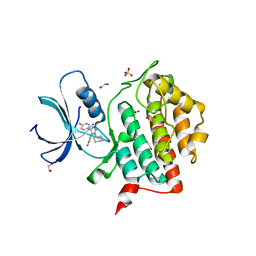 | | Crystal structure of CK1 delta in complex with VN725 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-cyclohexyl-5-(4-fluorophenyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Nemec, V, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3L5B
 
 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
7QR9
 
 | | Crystal structure of CK1 delta in complex with PK-09-82 | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(4-fluorophenyl)-3-(pyridin-4-ylmethyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QRB
 
 | | Crystal structure of CK1 delta in complex with PK-09-129 | | Descriptor: | 3-(dimethylamino)-~{N}-[4-[4-(4-fluorophenyl)-5-(1~{H}-pyrrolo[2,3-b]pyridin-4-yl)imidazol-1-yl]cyclohexyl]propane-1-sulfonamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
9H1F
 
 | |
8Q2M
 
 | | 18mer DNA mimic Foldamer with an Aliphatic linker in complex with Sac7d V26A/M29A protein | | Descriptor: | DNA mimic Foldamer, DNA-binding protein 7b | | Authors: | Deepak, D, Corvaglia, V, Wu, J, Huc, I. | | Deposit date: | 2023-08-02 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | DNA Mimic Foldamer Recognition of a Chromosomal Protein.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
1LFK
 
 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-11 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
4OL7
 
 | |
6QFA
 
 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6RM9
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RMB
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative mRNA splicing factor, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RMA
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RM8
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4RIE
 
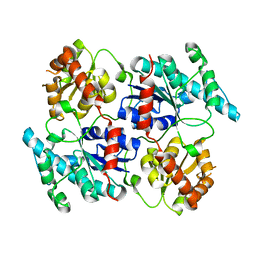 | | Landomycin Glycosyltransferase LanGT2 | | Descriptor: | Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIF
 
 | | Landomycin Glycosyltransferase LanGT2, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIG
 
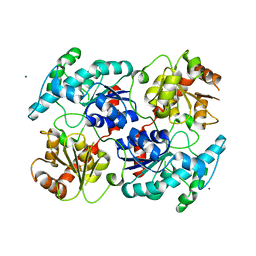 | | Chimeric Glycosyltransferase LanGT2S8Ac | | Descriptor: | Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIH
 
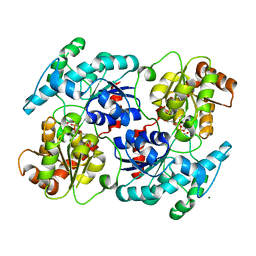 | | Chimeric Glycosyltransferase LanGT2S8Ac, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RII
 
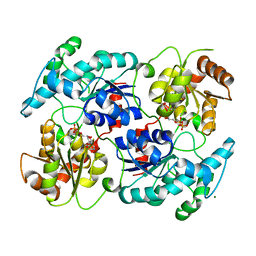 | | Chimeric Glycosyltransferase LanGT2S8Ac, TDP complex | | Descriptor: | Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2GHU
 
 | | Crystal structure of falcipain-2 from Plasmodium falciparum | | Descriptor: | falcipain 2 | | Authors: | Hogg, T, Nagarajan, K, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Characterization of Falcipain-2, a Hemoglobinase from the Malarial Parasite Plasmodium falciparum.
J.Biol.Chem., 281, 2006
|
|
6RMC
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative mRNA splicing factor, ... | | Authors: | Hamann, F, Neumann, P, Schmitt, A, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4PY7
 
 | | Crystal Structure of Fab 3.1 | | Descriptor: | antibody 3.1 heavy chain, antibody 3.1 light chain | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
