9FHV
 
 | |
9FHX
 
 | |
9FHY
 
 | |
9FHZ
 
 | |
9FI0
 
 | |
9FI2
 
 | |
9FI1
 
 | |
9FI3
 
 | |
9FI6
 
 | |
9QEL
 
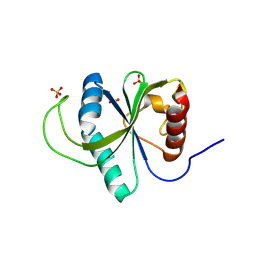 | |
9HDG
 
 | | The structure of the catalytic domain of AfAA11B from the filamentous fungus Aspergillus fumigatus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AA11 family lytic polysaccharide monooxygenase B, ASCORBIC ACID, ... | | Authors: | Hall, K, Roennekleiv, S.E, Gautieri, A, Skaali, R, Rieder, L, Englund, A.N, Landsem, E, Emrich-Mills, T, Ayuso-Fernandez, I, Golten, O, Kjendseth, A.R, Soerlie, M, Eijsink, V.G.H. | | Deposit date: | 2024-11-12 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Function Analysis of an Understudied Type of LPMO with Unique Redox Properties and Substrate Specificity.
Acs Catalysis, 15, 2025
|
|
9R7J
 
 | |
9EU3
 
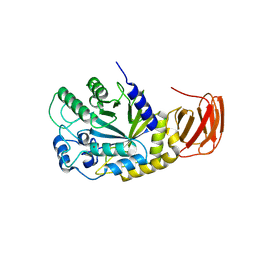 | | GH29A alpha-L-fucosidase | | Descriptor: | Alpha-L-fucosidase, CHLORIDE ION, ZINC ION, ... | | Authors: | Yang, Y.Y, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-03-27 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural elucidation and characterization of GH29A alpha-l-fucosidases and the effect of pH on their transglycosylation.
Febs J., 292, 2025
|
|
9FDH
 
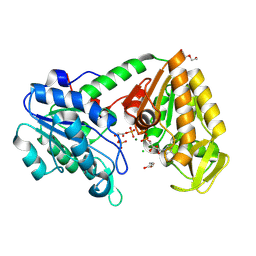 | | Closed Human phosphoglycerate kinase complex with BPG and ADP produced by cross-soaking a TSA crystal | | Descriptor: | 1,3-BISPHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Cliff, M.J, Waltho, J.P, Bowler, M.W, Baxter, N.J, Bisson, C, Blackburn, G.M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | The role of magnesium in catalysis by phosphoglycerate kinase
To be published
|
|
9FDN
 
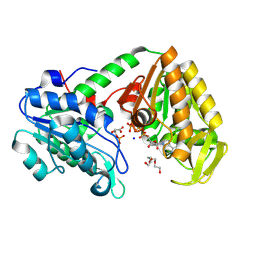 | | Human phosphoglycerate kinase in complex with ATP and 3PG formed by cross-soaking a TSA crystal | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Phosphoglycerate kinase 1, ... | | Authors: | Cliff, M.J, Waltho, J.P, Bowler, M.W, Baxter, N.J, Bisson, C, Blackburn, G.M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The role of magnesium in catalysis by phosphoglycerate kinase
To be published
|
|
9FFM
 
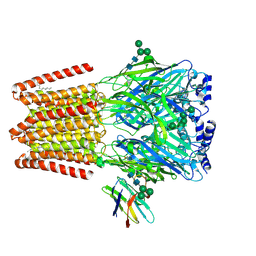 | |
9FF2
 
 | |
9FEZ
 
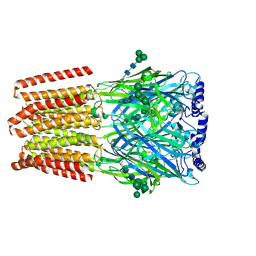 | |
9FFN
 
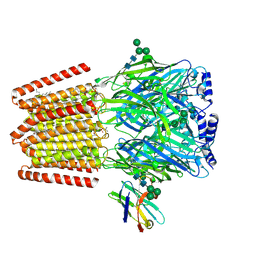 | |
9FUL
 
 | |
9FYU
 
 | |
9G8K
 
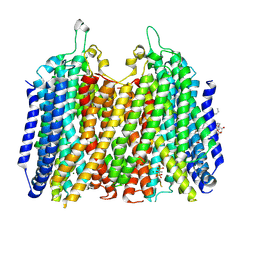 | | Structure of K+-dependent Na+-PPase from Thermotoga maritima in complex with Ca2+ and Etidronate | | Descriptor: | (1-hydroxyethane-1,1-diyl)bis(phosphonic acid), CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Vidilaseris, K, Liu, J, Goldman, A. | | Deposit date: | 2024-07-23 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Conformational dynamics and asymmetry in multimodal inhibition of membrane-bound pyrophosphatases
Elife, 2024
|
|
9GCA
 
 | |
9GFY
 
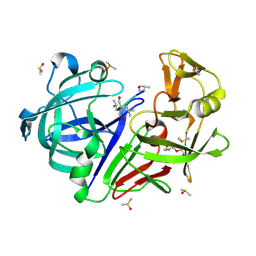 | | Endothiapepsin in complex with pepstatin soaked at pH 7.6 | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, PENTAETHYLENE GLYCOL, ... | | Authors: | Mueller, J.M, Glinca, S. | | Deposit date: | 2024-08-12 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Protonation Effects in Protein-Ligand Complexes - A Case Study of Endothiapepsin and Pepstatin A with Computational and Experimental Methods.
Chemmedchem, 20, 2025
|
|
9G6U
 
 | | p53-Y220C Core Domain Covalently Bound to 3,5-Dichloro-6-Ethylpyrazine-2-carbonitirle Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(chloranyl)-6-ethyl-pyrazine-2-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | SNAr Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes
Drug Des Devel Ther, 2025
|
|
