5H3K
 
 | |
5HEA
 
 | | CgT structure in hexamer | | Descriptor: | Putative glycosyltransferase (GalT1) | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2016-01-05 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A new helical binding domain mediates a glycosyltransferase activity of a bifunctional protein
To Be Published
|
|
8DL9
 
 | |
8DLB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | Descriptor: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
8DMD
 
 | |
5HEC
 
 | | CgT structure in dimer | | Descriptor: | Putative glycosyltransferase (GalT1) | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2016-01-05 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New Helical Binding Domain Mediates a Glycosyltransferase Activity of a Bifunctional Protein.
J.Biol.Chem., 291, 2016
|
|
4Z5V
 
 | |
5C94
 
 | | Infectious bronchitis virus nsp9 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-structural protein 9 | | Authors: | Chen, C, Dou, Y, Yang, H, Su, D. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9.
Protein Sci., 26, 2017
|
|
7LA6
 
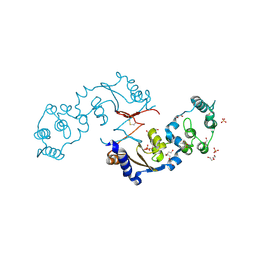 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, N239 deletion mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-05 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
4PHR
 
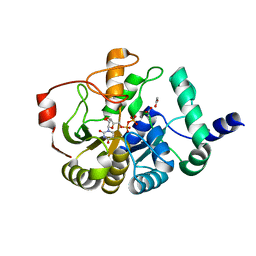 | | Domain of unknown function 1792 (DUF1792) with manganese | | Descriptor: | ACETATE ION, MANGANESE (II) ION, Putative glycosyltransferase (GalT1), ... | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold.
Nat Commun, 5, 2014
|
|
8XKO
 
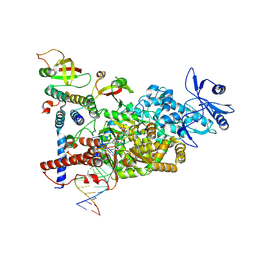 | | CryoEM structure of compound HNC-1664 bound with RdRP-RNA complex of SARS-CoV-2 | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, M, An, L, Hong, Y, Li, S, Zhang, K. | | Deposit date: | 2023-12-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | An adenosine analog shows high antiviral potency against coronavirus and arenavirus mainly through an unusual base pairing mode.
Nat Commun, 15, 2024
|
|
8XPO
 
 | |
8H1R
 
 | | Crystal structure of LptDE-YifL complex | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Surface lipoprotein sorting by crosstalk between Lpt and Lol pathways in gram-negative bacteria
Nat Commun, 16, 2025
|
|
8H1S
 
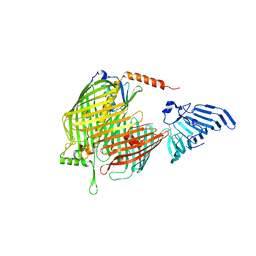 | | Crystal structure of apo-LptDE complex | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Surface lipoprotein sorting by crosstalk between Lpt and Lol pathways in gram-negative bacteria
Nat Commun, 16, 2025
|
|
7KB9
 
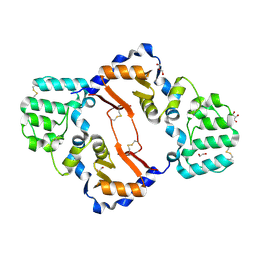 | | THE STRUCTURE OF A SENSOR DOMAIN OF A HISTIDINE KINASE (VxrA) FROM VIBRIO CHOLERAE O1 BIOVAR ELTOR STR. N16961, D238-T240 deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Sensor histidine kinase | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
8XPP
 
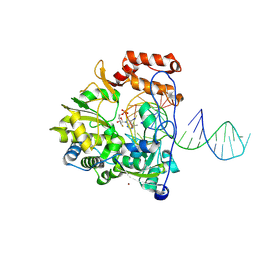 | |
4PFX
 
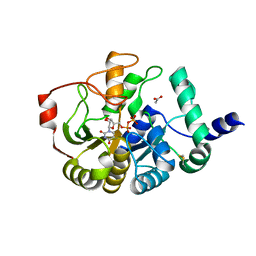 | |
4PHS
 
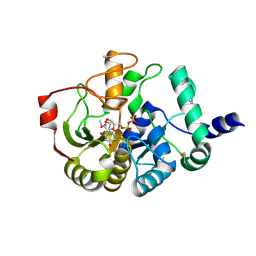 | |
2G17
 
 | |
5X0S
 
 | | Solution NMR structure of peptide toxin SsTx from Scolopendra subspinipes mutilans | | Descriptor: | SsTx | | Authors: | Wu, F, Luo, L, Qu, D, Zhang, L, Tian, C, Lai, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Centipedes subdue giant prey by blocking KCNQ channels
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7FEO
 
 | | Crystal structure of AtMBD5 MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 5, SULFATE ION | | Authors: | Zhou, M.Q, Wu, Z.B, Liu, K, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
