7MO3
 
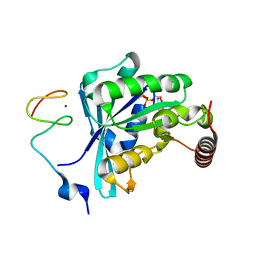 | | Crystal Structure of the ZnF3 of Nucleoporin NUP153 in complex with Ran-GDP, resolution 2.05 Angstrom | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MW0
 
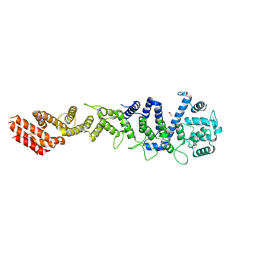 | | Crystal structure of Homo sapiens NUP93 solenoid (residues 174-819) | | Descriptor: | 1,2-ETHANEDIOL, Nuclear pore complex protein Nup93 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
170D
 
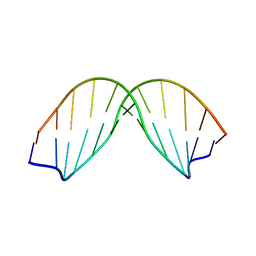 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | Descriptor: | DNA/RNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*T)-R(P*CAR)-D(P*GP*CP*G)-3') | | Authors: | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | Deposit date: | 1994-03-14 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
1ALE
 
 | |
171D
 
 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | Deposit date: | 1994-03-14 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
1CYF
 
 | |
1ALF
 
 | |
1CPG
 
 | |
1CPF
 
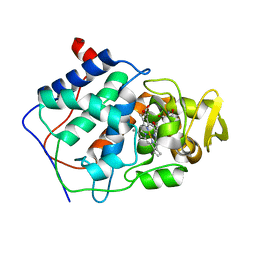 | | A CATION BINDING MOTIF STABILIZES THE COMPOUND I RADICAL OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miller, M.A, Han, G.W, Kraut, J. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cation binding motif stabilizes the compound I radical of cytochrome c peroxidase.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1CPD
 
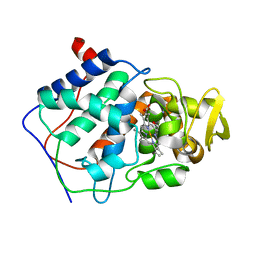 | |
1B8X
 
 | | GLUTATHIONE S-TRANSFERASE FUSED WITH THE NUCLEAR MATRIX TARGETING SIGNAL OF THE TRANSCRIPTION FACTOR AML-1 | | Descriptor: | PROTEIN (AML-1B) | | Authors: | Tang, L, Guo, B, Van Wijnen, A.J, Lian, J.B, Stein, J.L, Stein, G.S, Zhou, G.W. | | Deposit date: | 1999-02-03 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preliminary crystallographic study of glutathione S-transferase fused with the nuclear matrix targeting signal of the transcription factor AML-1/CBF-alpha2.
J.Struct.Biol., 123, 1998
|
|
1CPE
 
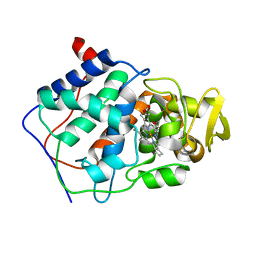 | |
1EWI
 
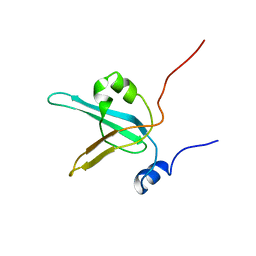 | | HUMAN REPLICATION PROTEIN A: GLOBAL FOLD OF THE N-TERMINAL RPA-70 DOMAIN REVEALS A BASIC CLEFT AND FLEXIBLE C-TERMINAL LINKER | | Descriptor: | REPLICATION PROTEIN A | | Authors: | Jacobs, D.M, Lipton, A.S, Isern, N.G, Daughdrill, G.W, Lowry, D.F, Gomes, X, Wold, M.S. | | Deposit date: | 2000-04-25 | | Release date: | 2000-05-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker.
J.Biomol.NMR, 14, 1999
|
|
1EM9
 
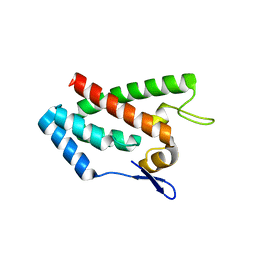 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: N-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27, MAGNESIUM ION | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-16 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
1E91
 
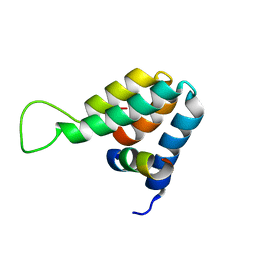 | | Structure of the complex of the Mad1-Sin3B interaction domains | | Descriptor: | MAD PROTEIN (MAX DIMERIZER), PAIRED AMPHIPATHIC HELIX PROTEIN SIN3B | | Authors: | Spronk, C.A.E.M, Tessari, M, Kaan, A.M, Jansen, J.F.A, Vermeulen, M, Stunnenberg, H.G, Vuister, G.W. | | Deposit date: | 2000-10-04 | | Release date: | 2000-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MAD1-Sin3B Interaction Involves a Novel Helical Fold
Nat.Struct.Biol., 7, 2000
|
|
1EAP
 
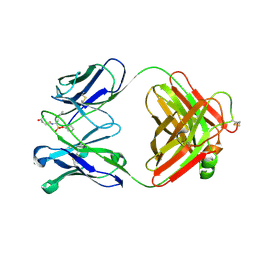 | | CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE | | Descriptor: | IGG2B-KAPPA 17E8 FAB (HEAVY CHAIN), IGG2B-KAPPA 17E8 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE | | Authors: | Zhou, G.W, Guo, J, Huang, W, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1994-08-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a catalytic antibody with a serine protease active site.
Science, 265, 1994
|
|
1EZE
 
 | | STRUCTURAL STUDIES OF A BABOON (PAPIO SP.) PLASMA PROTEIN INHIBITOR OF CHOLESTERYL ESTER TRANSFERASE. | | Descriptor: | CHOLESTERYL ESTER TRANSFERASE INHIBITOR PROTEIN | | Authors: | Buchko, G.W, Rozek, A, Kanda, P, Kennedy, M.A, Cushley, R.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a baboon (Papio sp.) plasma protein inhibitor of cholesteryl ester transferase.
Protein Sci., 9, 2000
|
|
6SD9
 
 | | Crystal structure of wild-type cMET bound by foretinib | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDC
 
 | | Crystal structure of D1228V cMET bound by foretinib | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDD
 
 | | Crystal structure of D1228V cMET bound by BMS-777607 | | Descriptor: | GLYCEROL, Hepatocyte growth factor receptor, N-{4-[(2-amino-3-chloropyridin-4-yl)oxy]-3-fluorophenyl}-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDE
 
 | |
1FK2
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH MYRISTIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, MYRISTIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK7
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH RICINOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN, RICINOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK3
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK0
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-08 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
