8AKM
 
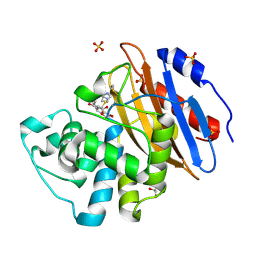 | | Acyl-enzyme complex of ertapenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, Ertapenem, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKK
 
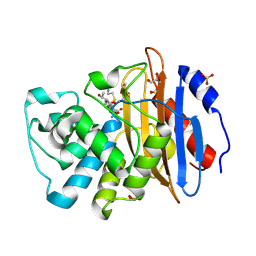 | | Acyl-enzyme complex of imipenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKJ
 
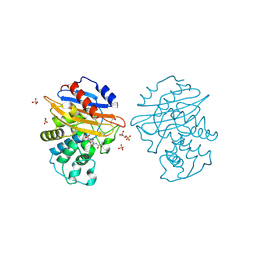 | | Acyl-enzyme complex of cephalothin bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKL
 
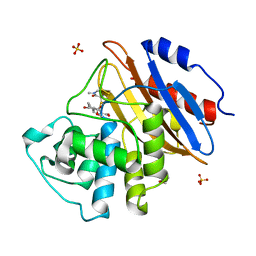 | | Acyl-enzyme complex of meropenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2S,3R,4R)-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl-3-methyl-2-[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
9BD5
 
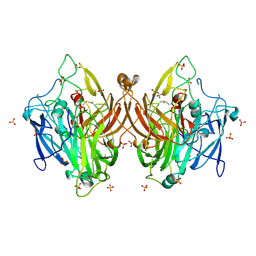 | | Laccase from Bacillus licheniformis | | Descriptor: | COPPER (II) ION, SULFATE ION, Spore coat protein A | | Authors: | Habib, M.H, Smith, T.J. | | Deposit date: | 2024-04-11 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polymerization potential of a bacterial CotA-laccase for beta-naphthol: enzyme structure and comprehensive polymer characterization.
Front Microbiol, 15, 2024
|
|
9B8J
 
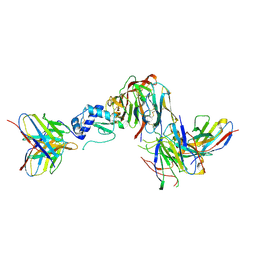 | | GP38-GnH-DS-Gc in the pre-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-36125 heavy chain, ADI-36125 light chain, ... | | Authors: | McFadden, E, McLellan, J.S. | | Deposit date: | 2024-03-30 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Engineering and structures of Crimean-Congo hemorrhagic fever virus glycoprotein complexes.
Cell, 188, 2025
|
|
9CSF
 
 | |
9CSH
 
 | |
9C97
 
 | |
9CMR
 
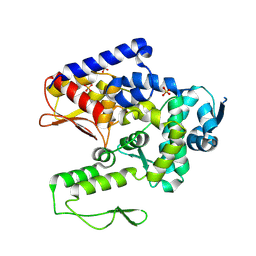 | | Saccharomyces cerevisiae Tom1 HECT domain | | Descriptor: | E3 ubiquitin-protein ligase TOM1, SULFATE ION | | Authors: | Schubert, H.L, Hill, C.P. | | Deposit date: | 2024-07-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tom1p ubiquitin ligase structure, interaction with Spt6p, and function in maintaining normal transcript levels and the stability of chromatin in promoters.
To Be Published
|
|
9E3B
 
 | | Cryo-EM structure of PRMT5/WDR77 in complex with 6S complex | | Descriptor: | Methylosome protein WDR77, Methylosome subunit pICln, Protein arginine N-methyltransferase 5, ... | | Authors: | Jin, C.Y, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2024-10-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Substrate adaptors are flexible tethering modules that enhance substrate methylation by the arginine methyltransferase PRMT5.
J.Biol.Chem., 301, 2025
|
|
9DGZ
 
 | |
9DH0
 
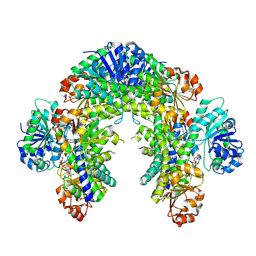 | | The Cryo-EM structure of recombinantly expressed hUGDH in complex with UDP-4-keto-xylose | | Descriptor: | (2R,3R,4R)-3,4-dihydroxy-5-oxooxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), UDP-glucose 6-dehydrogenase | | Authors: | Kadirvelraj, R, Walsh Jr, R.M, Wood, Z.W. | | Deposit date: | 2024-09-03 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM Structure of Recombinantly Expressed hUGDH Unveils a Hidden, Alternative Allosteric Inhibitor.
Biochemistry, 64, 2025
|
|
9EGW
 
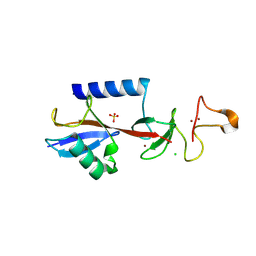 | |
9EGV
 
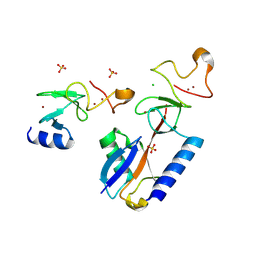 | | HOIL-1 RING2 domain bound to ubiquitin | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, RanBP-type and C3HC4-type zinc finger-containing protein 1, ... | | Authors: | Wang, X.S, Lechtenberg, B.C. | | Deposit date: | 2024-11-21 | | Release date: | 2024-12-11 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The RBR E3 ubiquitin ligase HOIL-1 can ubiquitinate diverse non-protein substrates in vitro.
Life Sci Alliance, 8, 2025
|
|
9EC2
 
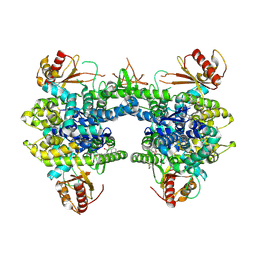 | | Crystal structure of SAMHD1 dimer bound to an inhibitor obtained from high-throughput chemical tethering to the guanine antiviral acyclovir | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, N-[5-({2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl}amino)-5-oxopentyl]-4,7-dibromo-3-hydroxynaphthalene-2-carboxamide | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Lopez-Rovira, L.M, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2024-11-13 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Inhibitors of SAMHD1 Obtained from Chemical Tethering to the Guanine Antiviral Acyclovir.
Biochemistry, 64, 2025
|
|
9DYB
 
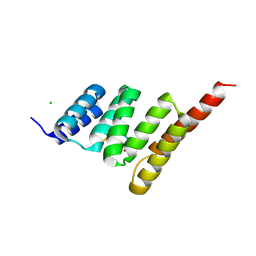 | |
9DYA
 
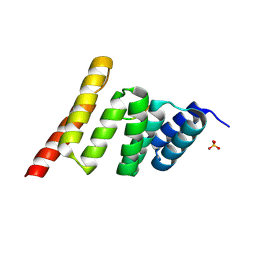 | | CHIP-TPR in complex with the Hsp70 tail | | Descriptor: | E3 ubiquitin-protein ligase CHIP, Heat shock 70 kDa protein 1A, SULFATE ION | | Authors: | Zhang, H, Nix, J.C, Page, R.C. | | Deposit date: | 2024-10-13 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Phosphorylation-State Modulated Binding of HSP70: Structural Insights and Compensatory Protein Engineering.
Biorxiv, 2025
|
|
9E6S
 
 | |
9E6T
 
 | |
9C12
 
 | | Leukotriene C4-bound Multidrug Resistance-associated Protein 2 (MRP2) | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-( 2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, ATP-binding cassette sub-family C member 2, CHOLESTEROL, ... | | Authors: | Koide, E, Chen, J. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis for the transport and regulation mechanism of the multidrug resistance-associated protein 2.
Nat Commun, 16, 2025
|
|
9D7I
 
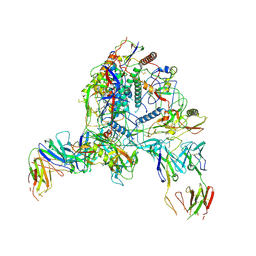 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 2 CH103 KN Fabs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab light chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7H
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 1 CH103 KN Fab bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab light chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7G
 
 | | BG505 DS-SOSIP.664 apo structure from the CH103 KN cryo-EM dataset | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface protein gp120, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
9D7O
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 1 CH103 Fab bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab heavy chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 2025
|
|
