3A6D
 
 | | Creatininase complexed with 1-methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6E
 
 | | W174F mutant creatininase, type I | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6L
 
 | | E122Q mutant creatininase, Zn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6H
 
 | | W154A mutant creatininase | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6F
 
 | | W174F mutant creatininase, Type II | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6G
 
 | | W154F mutant creatininase | | Descriptor: | Creatinine amidohydrolase, MANGANESE (II) ION, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
5YZ7
 
 | | Crystal structure of OsD14 in complex with D-ring-opened 7'-carba-4BD | | Descriptor: | (2Z,4S)-5-(4-bromophenyl)-4-hydroxy-2-methylpent-2-enoic acid, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Jiang, K, Xu, Y, Miyakawa, T, Asami, T, Tanokura, M. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Rationally Designed Strigolactone Analogs as Antagonists of the D14 Receptor.
Plant Cell Physiol., 59, 2018
|
|
3AGK
 
 | | Crystal structure of archaeal translation termination factor, aRF1 | | Descriptor: | Peptide chain release factor subunit 1 | | Authors: | Kobayashi, K, Kikuno, I, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Omnipotent role of archaeal elongation factor 1 alpha (EF1{alpha}) in translational elongation and termination, and quality control of protein synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5XEJ
 
 | |
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVB
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVC
 
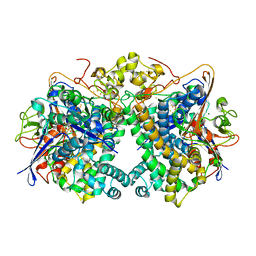 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
3W3E
 
 | |
1VGO
 
 | | Crystal Structure of Archaerhodopsin-2 | | Descriptor: | Archaerhodopsin 2, RETINAL, SULFATE ION, ... | | Authors: | Yoshimura, K, Enami, N, Murakami, M, Okumura, H, Ihara, K, Kouyama, T. | | Deposit date: | 2004-04-28 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps
J.Mol.Biol., 358, 2006
|
|
3WP3
 
 | |
2DXS
 
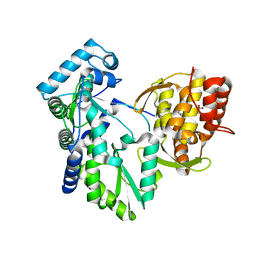 | | Crystal structure of HCV NS5B RNA polymerase complexed with a tetracyclic inhibitor | | Descriptor: | Genome polyprotein, N-[(13-CYCLOHEXYL-6,7-DIHYDROINDOLO[1,2-D][1,4]BENZOXAZEPIN-10-YL)CARBONYL]-2-METHYL-L-ALANINE | | Authors: | Adachi, T, Tsuruha, J, Doi, S, Murase, K, Ikegashira, K, Watanabe, S, Uehara, K, Orita, T, Nomura, A, Kamada, M. | | Deposit date: | 2006-08-30 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Conformationally Constrained Tetracyclic Compounds as Potent Hepatitis C Virus NS5B RNA Polymerase Inhibitors
J.Med.Chem., 49, 2006
|
|
2Z55
 
 | |
3AXZ
 
 | | Crystal structure of Haemophilus influenzae TrmD in complex with adenosine | | Descriptor: | ADENOSINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Yoshida, K, Goto-Ito, S, Ito, T, Hou, Y.M, Yokoyama, S. | | Deposit date: | 2011-04-21 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Differentiating analogous tRNA methyltransferases by fragments of the methyl donor.
Rna, 17, 2011
|
|
6E8C
 
 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | Authors: | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
418D
 
 | |
7R9C
 
 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7MC5
 
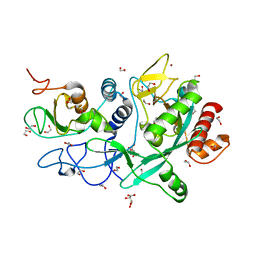 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC6
 
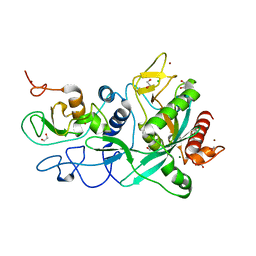 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3K55
 
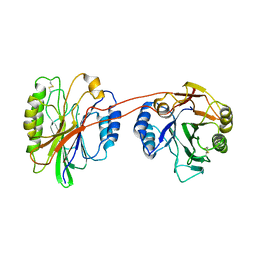 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | Descriptor: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-10-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
