5M5G
 
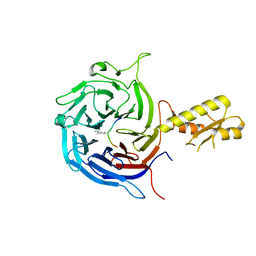 | | Crystal structure of the Chaetomium Thermophilum polycomb repressive complex 2 (PRC2) | | Descriptor: | Fragment from molecular 2 (region containing putative polycomb protein Suz12), HISTONE H3 11-Mer peptide, Putative uncharacterized protein, ... | | Authors: | Zhang, Y, Justin, N, Wilson, J, Gamblin, S. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Science, 354, 2016
|
|
5E4K
 
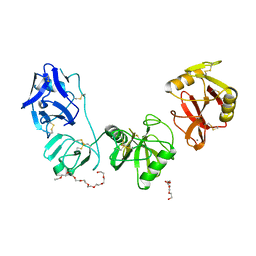 | | Structure of ligand binding region of uPARAP at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
5EW6
 
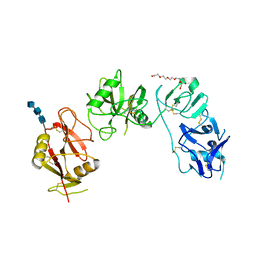 | | Structure of ligand binding region of uPARAP at pH 7.4 without calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
8Y6F
 
 | |
4OVA
 
 | |
4XXB
 
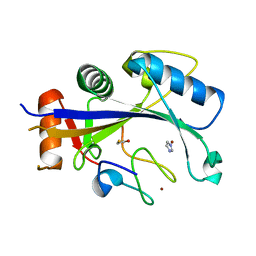 | | Crystal structure of human MDM2-RPL11 | | Descriptor: | 60S ribosomal protein L11, BETA-MERCAPTOETHANOL, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Zheng, J, Chen, Z. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation
Genes Dev., 29, 2015
|
|
8K9V
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 3 (ADKI3) | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9X
 
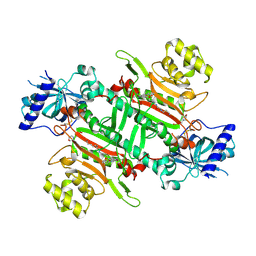 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 5 (ADKI5) | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-[3-[2-[[4-[(2,5-dimethoxyphenyl)amino]-1,3,5-triazin-2-yl]amino]phenyl]sulfonylpropyl]hexanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lysine--tRNA ligase | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9S
 
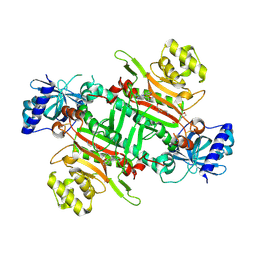 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 1 (ADKI1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9U
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 2 (ADKI2) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9W
 
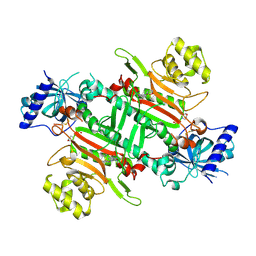 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 4 (ADKI4) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lysine--tRNA ligase, ~{N}2-(2-methoxyphenyl)-~{N}4-(2-propan-2-ylsulfonylphenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
4ZSZ
 
 | |
4ZSV
 
 | |
4ZSX
 
 | |
5ZHH
 
 | |
6AZR
 
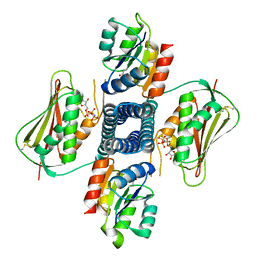 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
8YJN
 
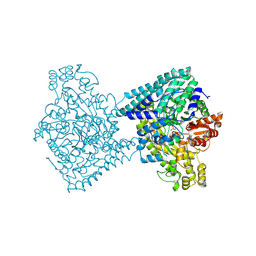 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8YJO
 
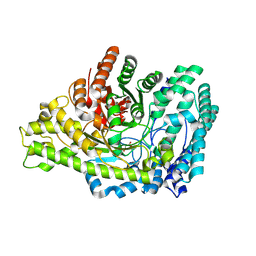 | | Structure of E. coli glycyl radical enzyme PflD with bound malonate | | Descriptor: | MALONATE ION, Probable dehydratase PflD | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
3IEI
 
 | |
4OLS
 
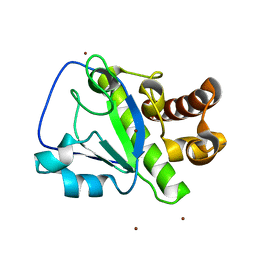 | | The amidase-2 domain of LysGH15 | | Descriptor: | Endolysin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
8I2J
 
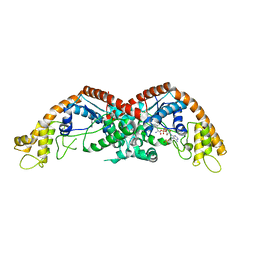 | |
8I1Y
 
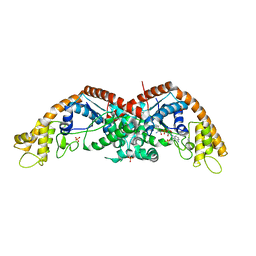 | | The structure of E. coli TrpRS bound with a chemical fragment | | Descriptor: | 5-ethanoylthiophene-2-carbonitrile, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I27
 
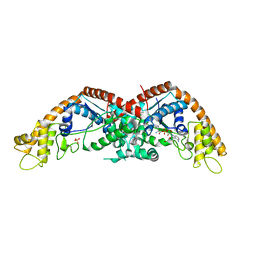 | |
8I2A
 
 | |
8I4I
 
 | |
