7DF9
 
 | | Crystal of Arrestin2-V2Rpp-1-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DF8
 
 | | full length hNPC1L1-Apo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, M, Sun, S, Sui, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into the mechanism of human NPC1L1-mediated cholesterol uptake.
Sci Adv, 7, 2021
|
|
7DFA
 
 | | Crystal of Arrestin2-V2Rpp-4-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFC
 
 | | Crystal of Arrestin2-V2Rpp-3-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFW
 
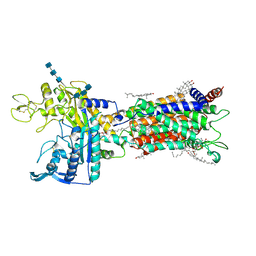 | | Cryo_EM structure of delta N-NPC1L1-CLR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, M, Sun, S. | | Deposit date: | 2020-11-10 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into the mechanism of human NPC1L1-mediated cholesterol uptake.
Sci Adv, 7, 2021
|
|
7DFZ
 
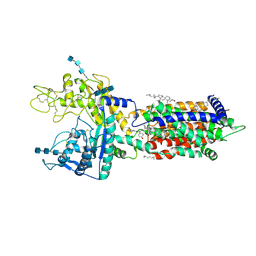 | | Cryo_EM structure of delta N-NPC1L1-EZE | | Descriptor: | (3~{R},4~{S})-1-(4-fluorophenyl)-3-[(3~{S})-3-(4-fluorophenyl)-3-oxidanyl-propyl]-4-(4-hydroxyphenyl)azetidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, M, Sun, S. | | Deposit date: | 2020-11-10 | | Release date: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into the mechanism of human NPC1L1-mediated cholesterol uptake.
Sci Adv, 7, 2021
|
|
7DJJ
 
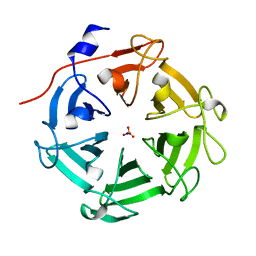 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | Descriptor: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69806433 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJM
 
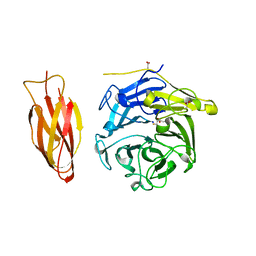 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70000112 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
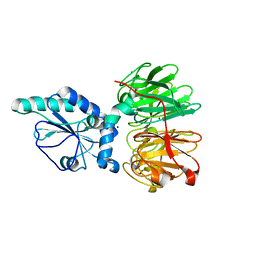 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.80145121 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJL
 
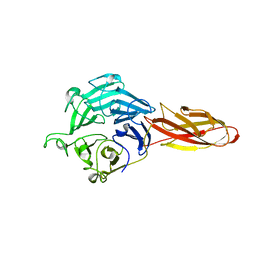 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96077824 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7BZJ
 
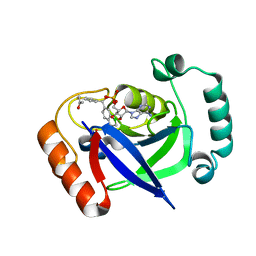 | | The Discovery of Benzhydrol-Oxaborole Hybrid Derivatives as Leucyl-tRNA Synthetase Inhibitors | | Descriptor: | Leucine--tRNA ligase, [(1~{R},5~{R},6~{S},8~{R})-8-(6-aminopurin-9-yl)-4'-[(~{R})-oxidanyl-[4-(2-oxidanylidenepropylsulfanyl)phenyl]methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,7'-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-6-yl]methoxy-tris(oxidanyl)phosphanium | | Authors: | Liu, R.J, Li, H, Wang, E.D, Zhou, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of benzhydrol-oxaborole derivatives as Streptococcus pneumoniae leucyl-tRNA synthetase inhibitors.
Bioorg.Med.Chem., 29, 2021
|
|
7E5W
 
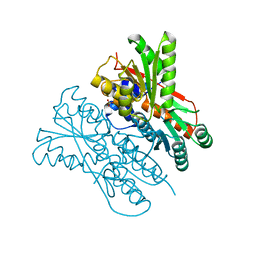 | | The structure of CcpA from Staphylococcus aureus | | Descriptor: | Catabolite control protein A, SULFATE ION | | Authors: | Yu, G, Wei, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Regulation of DNA-binding activity of the Staphylococcus aureus catabolite control protein A by copper (II)-mediated oxidation.
J.Biol.Chem., 298, 2022
|
|
7EEA
 
 | |
7EEQ
 
 | | Cyanophage Pam1 tail machine | | Descriptor: | Needle head proteins, Tailspike head-binding domain | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEP
 
 | | Cyanophage Pam1 portal-adaptor complex | | Descriptor: | Pam1 adaptor proteins, Pam1 portal proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEL
 
 | | Cyanophage Pam1 capsid asymmetric unit | | Descriptor: | Cement (decoration) proteins, Major capsid proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7E8T
 
 | | Monomer of Ypt32-TRAPPII | | Descriptor: | GTP-binding protein YPT32/YPT11, TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-02 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E2C
 
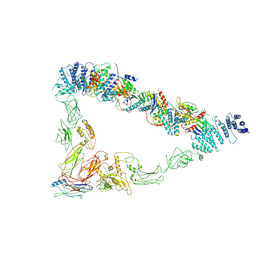 | | Monomer of TRAPPII (open) | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Sui, S.F, Sun, S, Mi, C.C. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E94
 
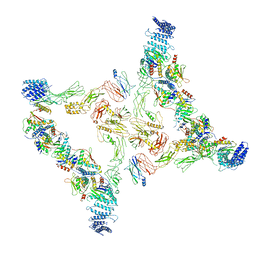 | | Intact TRAPPII (State II) | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E93
 
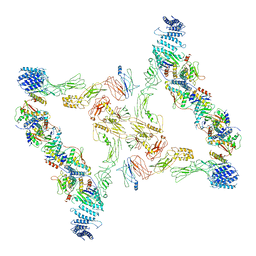 | | Intact TRAPPII (state III). | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.54 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7EA3
 
 | | Intact Ypt32-TRAPPII (dimer). | | Descriptor: | GTP-binding protein YPT32/YPT11, TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E8S
 
 | | Intact TRAPPII (state I). | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-02 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E2D
 
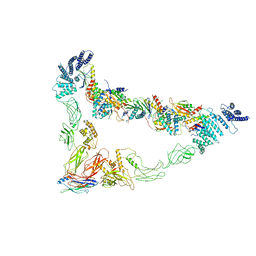 | | Monomer of TRAPPII (Closed) | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Sui, S.F, Sun, S, Mi, C.C. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E6V
 
 | | The crystal structure of foot-and-mouth disease virus(FMDV) 2C protein 97-318aa | | Descriptor: | ACETATE ION, Protein 2C | | Authors: | Zhang, C, Wojdyla, J.A, Qin, B, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2021-02-24 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | An anti-picornaviral strategy based on the crystal structure of foot-and-mouth disease virus 2C protein.
Cell Rep, 40, 2022
|
|
7VC9
 
 | | Tom20 subunits | | Descriptor: | Mitochondrial import receptor subunit TOM20 homolog | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
