300D
 
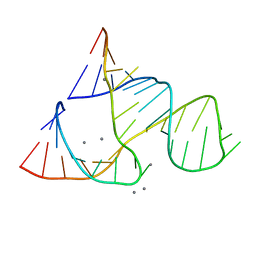 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MN(II)-SOAKED | | Descriptor: | MANGANESE (II) ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
301D
 
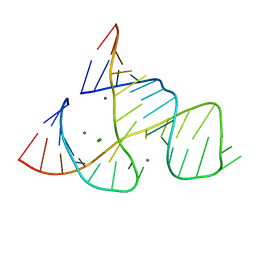 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MG(II)-SOAKED | | Descriptor: | MAGNESIUM ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
3AEH
 
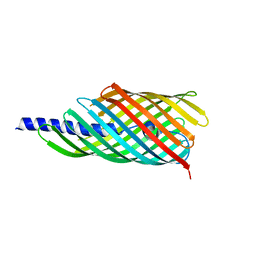 | |
5F73
 
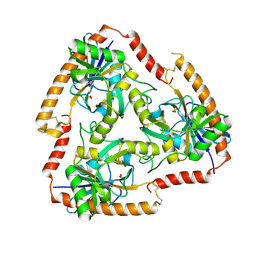 | | Crystal structure of Mutant S12T of Adenosine/Methylthioadenosine Phosphorylase in APO form | | Descriptor: | Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
6DQ6
 
 | |
6DQF
 
 | |
5F77
 
 | | Crystal structure of Mutant S12T of adenosine/Methylthioadenosine phosphorylase from Schistosoma mansoni in complex with Adenine | | Descriptor: | ADENINE, Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
6DQA
 
 | | Linked KDM5A JMJ Domain Bound to Inhibitor N70 i.e.[2-((3-aminophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-(3-aminophenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
5F76
 
 | | Crystal structure of Mutant S12T of Adenosine/Methylthioadenosine Phosphorylase from Schistosoma mansoni in complex with Methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
6DQC
 
 | |
6DQ7
 
 | |
6DQ9
 
 | | Linked KDM5A JMJ Domain Bound to the Covalent Inhibitor N69 i.e. [2-((3-acrylamidophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-[3-(acryloylamino)phenyl][2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQD
 
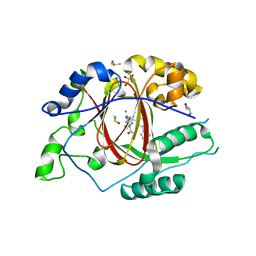 | |
6DQ5
 
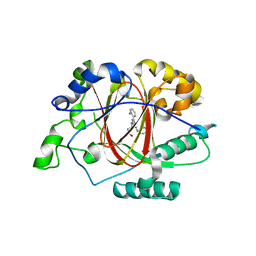 | |
6DQE
 
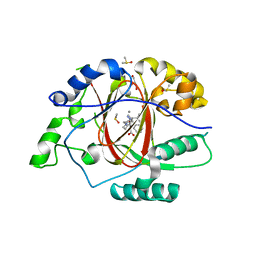 | |
4EVN
 
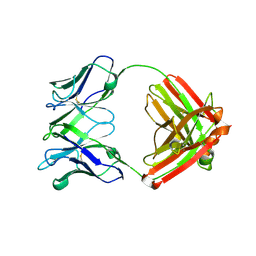 | |
4G1I
 
 | |
3I4R
 
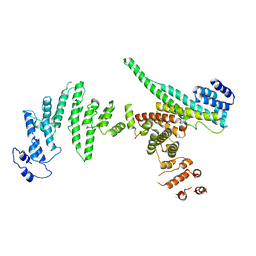 | | Nup107(aa658-925)/Nup133(aa517-1156) complex, H.sapiens | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133 | | Authors: | Whittle, J.R.R, Schwartz, T.U. | | Deposit date: | 2009-07-02 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
J.Biol.Chem., 284, 2009
|
|
3I5P
 
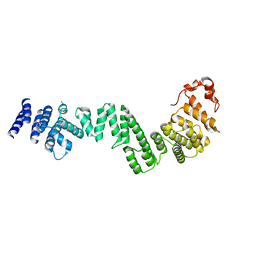 | | Nup170(aa979-1502), S.cerevisiae | | Descriptor: | Nucleoporin NUP170 | | Authors: | Whittle, J.R.R, Schwartz, T.U. | | Deposit date: | 2009-07-06 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
J.Biol.Chem., 284, 2009
|
|
5CUK
 
 | | Crystal structure of the PscP SS domain | | Descriptor: | GLYCEROL, Ruler protein | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
3I5Q
 
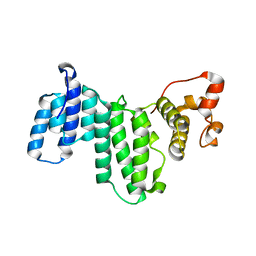 | | Nup170(aa1253-1502) at 2.2 A, S.cerevisiae | | Descriptor: | Nucleoporin NUP170 | | Authors: | Whittle, J.R.R, Schwartz, T.U. | | Deposit date: | 2009-07-06 | | Release date: | 2009-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
J.Biol.Chem., 284, 2009
|
|
5CUL
 
 | | crystal structure of the PscU C-terminal domain | | Descriptor: | Translocation protein in type III secretion | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
5F78
 
 | | Crystal structure of Mutant N87T of adenosine/Methylthioadenosine phosphorylase from Schistosoma mansoni in APO form | | Descriptor: | Methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R.S, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8518 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
6DQ4
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
4G08
 
 | |
