6L4S
 
 | |
5XYY
 
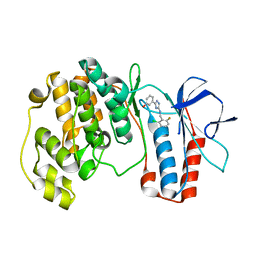 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | 3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-4H-1,2,4-triazol-3-yl)phenol, Mitogen-activated protein kinase 14 | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
8ESH
 
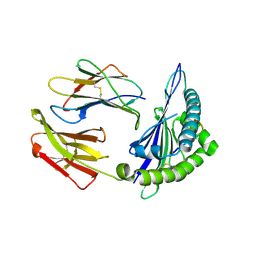 | | Structure of chimeric HLA-A*02:01 bound to CMV peptide | | Descriptor: | Beta-2-microglobulin, CMV peptide, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
8ERX
 
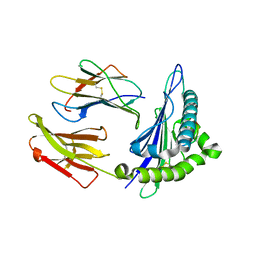 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
7XDQ
 
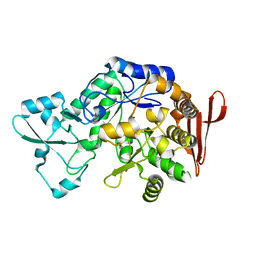 | | Crystal structure of a glucosylglycerol phosphorylase mutant from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase, LITHIUM ION, beta-D-glucopyranose | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
7XDR
 
 | | Crystal structure of a glucosylglycerol phosphorylase from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
3AL4
 
 | | Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Zhang, W, Qi, J.X, Shi, Y, Li, Q, Yan, J.H, Gao, G.F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus
Protein Cell, 1, 2010
|
|
5GTD
 
 | |
7YMN
 
 | | Cryo-EM structure of in vitro PHF fibril | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Li, X, Liu, C. | | Deposit date: | 2022-07-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Subtle change of fibrillation condition leads to substantial alteration of recombinant Tau fibril structure.
Iscience, 25, 2022
|
|
7YPG
 
 | |
6XIE
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 77 | | Descriptor: | GLYCEROL, Peptide 77, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6XIC
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 40 | | Descriptor: | GLYCEROL, Peptide 40, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
7VUN
 
 | | Design, modification, evaluation and cocrystal studies of novel phthalimides regulating PD-1/PD-L1 interaction | | Descriptor: | (2~{S},3~{S})-2-[[6-[(3-cyanophenyl)methoxy]-2-(2-methyl-3-phenyl-phenyl)-1,3-bis(oxidanylidene)isoindol-5-yl]methylamino]-3-oxidanyl-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Sun, C.L, Chen, M.R, Yang, P, Xiao, Y.B. | | Deposit date: | 2021-11-03 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Novel phthalimides regulating PD-1/PD-L1 interaction as potential immunotherapy agents.
Acta Pharm Sin B, 12, 2022
|
|
6XIF
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 83 | | Descriptor: | GLYCEROL, Peptide 83, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6XID
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 51 | | Descriptor: | GLYCEROL, Peptide 51, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6XIB
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 30 | | Descriptor: | GLYCEROL, Peptide 30, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
7WL2
 
 | |
7WLE
 
 | |
7WL9
 
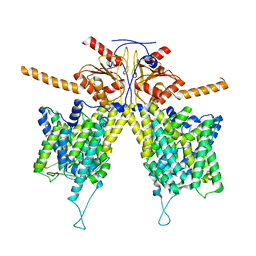 | | Mouse Pendrin in chloride and bicarbonate in asymmetric state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-12 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WLA
 
 | |
7WK7
 
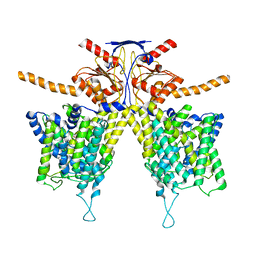 | | Mouse Pendrin bound bicarbonate in inward state | | Descriptor: | BICARBONATE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-05-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WK1
 
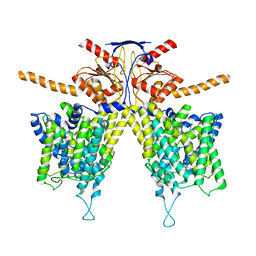 | | Mouse Pendrin bound chloride in inward state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WL7
 
 | |
7WL8
 
 | |
7WLB
 
 | |
