8IYO
 
 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
8IYM
 
 | | Crystal structure of a protein acetyltransferase, HP0935 | | Descriptor: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
5NCK
 
 | |
8J09
 
 | | Crystal structure of the Sld3 Cdc45-binding-domain, in complex with Cdc45 | | Descriptor: | Cell division control protein 45, DNA replication regulator SLD3 | | Authors: | Li, H, Ishizaka, I, Kato, K, Sun, X, Muramatsu, S, Itou, H, Ose, T, Araki, H, Yao, M. | | Deposit date: | 2023-04-10 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into Cdc45 recruitment by Sld7-Sld3 for CMG complex formation
Elife, 2024
|
|
5ZKA
 
 | | Crystal structure of N-acetylneuraminate lyase from Fusobacterium nucleatum complexed with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, N-acetylneuraminate lyase, TRIETHYLENE GLYCOL | | Authors: | Kumar, J.P, Rao, H, Nayak, V, Ramaswamy, S. | | Deposit date: | 2018-03-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures and kinetics of N-acetylneuraminate lyase from Fusobacterium nucleatum
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5VJ5
 
 | | Horse Liver Alcohol Dehydrogenase Complexed with 1,10-Phenanthroline | | Descriptor: | 1,10-PHENANTHROLINE, Alcohol dehydrogenase E chain, ZINC ION | | Authors: | Plapp, B.V, Baskar Raj, S, Ramaswamy, S. | | Deposit date: | 2017-04-18 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Horse Liver Alcohol Dehydrogenase: Zinc Coordination and Catalysis.
Biochemistry, 56, 2017
|
|
6AC7
 
 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
4QVK
 
 | | Apo-crystal structure of Podospora anserina methyltransferase PaMTH1 | | Descriptor: | 1,2-ETHANEDIOL, PaMTH1 Methyltransferase | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2014-07-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
6EBK
 
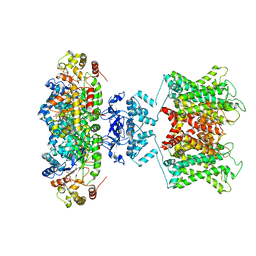 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6EBL
 
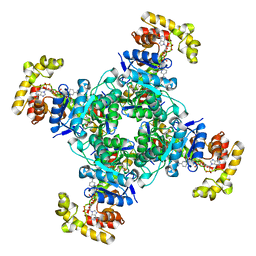 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6FKX
 
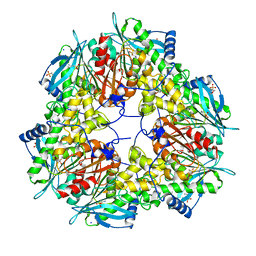 | | Crystal structure of an acetyl xylan esterase from a desert metagenome | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetyl xylan esterase, FORMIC ACID, ... | | Authors: | Adesioye, F.A, Makhalanyane, T.P, Vikram, S, Sewell, B.T, Schubert, W, Cowan, D.A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Characterization and Directed Evolution of a Novel Acetyl Xylan Esterase Reveals Thermostability Determinants of the Carbohydrate Esterase 7 Family.
Appl. Environ. Microbiol., 84, 2018
|
|
6VYF
 
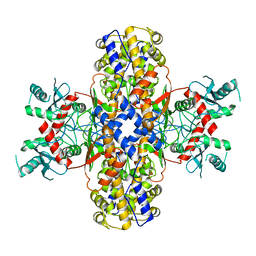 | | Cryo-EM structure of Plasmodium vivax hexokinase (Open state) | | Descriptor: | Phosphotransferase | | Authors: | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
6VYG
 
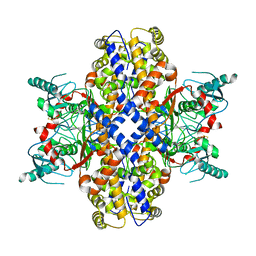 | | Cryo-EM structure of Plasmodium vivax hexokinase (Closed state) | | Descriptor: | Phosphotransferase | | Authors: | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
5VKR
 
 | |
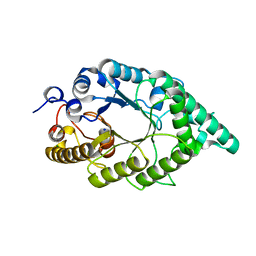
5EBA
 
 | |
5EB8
 
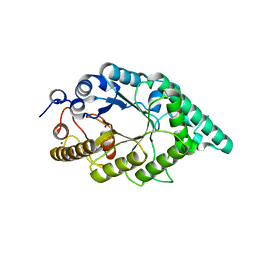 | |
5EFF
 
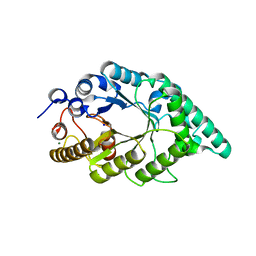 | | Crystal structure of an aromatic mutant (F4A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Beta-xylanase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of an aromatic mutant (F4A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27
To Be Published
|
|
5EFD
 
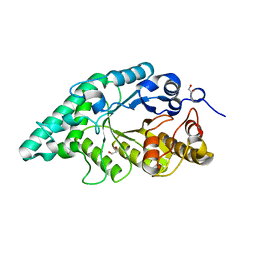 | | Crystal structure of a surface pocket creating mutant (W6A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CHLORIDE ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
5XC0
 
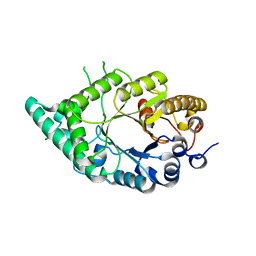 | |
5XOZ
 
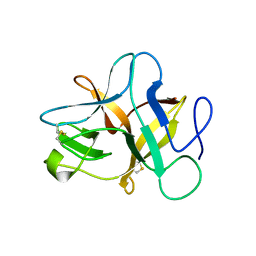 | |
7KD8
 
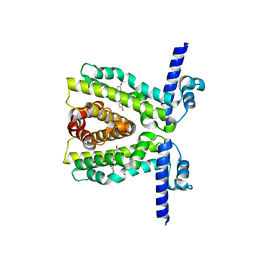 | | TtgR C137I I141W M167L F168Y mutant in complex with resveratrol | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1A
 
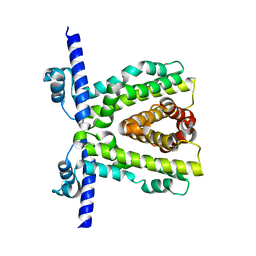 | | TtgR quadruple mutant (C137I I141W M167L F168Y) | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1C
 
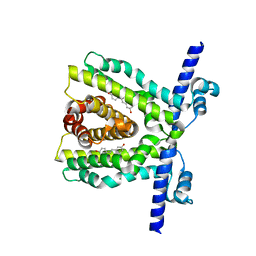 | | TtgR in complex with resveratrol | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7L8K
 
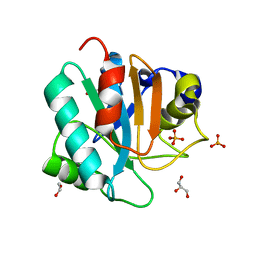 | | Crystal structure of human GPX4-U46C | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
7L8Q
 
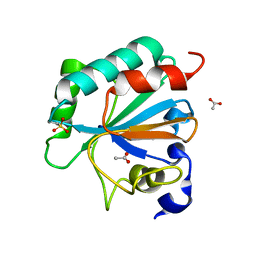 | | Crystal structure of human GPX4-U46C with oxidized Cys-46 | | Descriptor: | ACETATE ION, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
