4OZW
 
 | |
3T5S
 
 | |
3HGU
 
 | |
3PSI
 
 | |
4NX4
 
 | | Re-refinement of CAP-1 HIV-CA complex | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Lang, P.T, Holton, J.M, Fraser, J.S, Alber, T. | | Deposit date: | 2013-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein structural ensembles are revealed by redefining X-ray electron density noise.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3PSJ
 
 | |
3N0Y
 
 | | Adenylate cyclase class IV with active site ligand APC | | Descriptor: | Adenylate cyclase 2, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
3UO1
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, 64-3-7 L CHAIN, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-11-16 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3N0Z
 
 | |
3N10
 
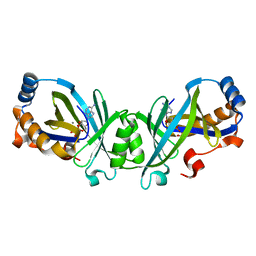 | | Product complex of adenylate cyclase class IV | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Adenylate cyclase 2, MANGANESE (II) ION, ... | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
1OMO
 
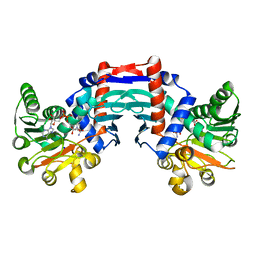 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
3PSK
 
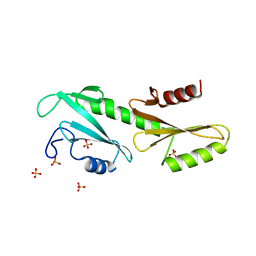 | |
4A2U
 
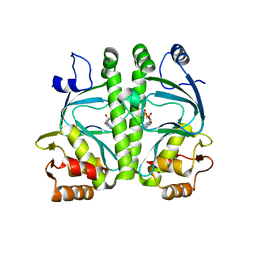 | | CRP(CAP) from Myco. Tuberculosis, with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY CRP/ FNR-FAMILY) | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2011-09-28 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | A New Crystal Form of Crp from M. Tuberculosis and Conformational Effects of Crystal Packing
To be Published
|
|
3UYR
 
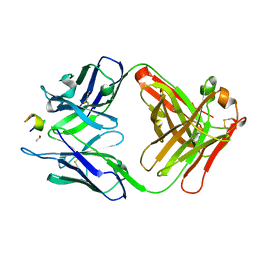 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, H-2 class I histocompatibility antigen, L-D alpha chain, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-12-06 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3H78
 
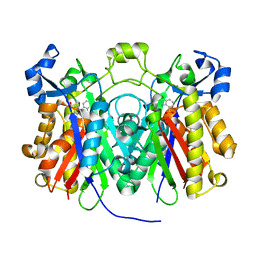 | | Crystal structure of Pseudomonas aeruginosa PqsD C112A mutant in complex with anthranilic acid | | Descriptor: | 2-AMINOBENZOIC ACID, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3H76
 
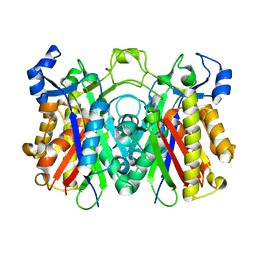 | | Crystal structure of PqsD, a key enzyme in Pseudomonas aeruginosa quinolone signal biosynthesis pathway | | Descriptor: | PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3V4U
 
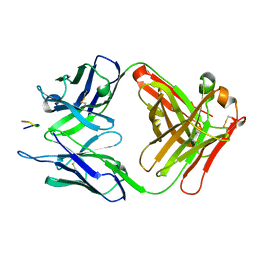 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, 64-3-7 L CHAIN, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
3JRM
 
 | |
3JSE
 
 | |
3PSF
 
 | |
4OZV
 
 | | Crystal Structure of the periplasmic alginate lyase AlgL | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid | | Authors: | Howell, P.L, Wolfram, F, Robinson, H, Arora, K. | | Deposit date: | 2014-02-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | The Pseudomonas aeruginosa homeostasis enzyme AlgL clears the periplasmic space of accumulated alginate during polymer biosynthesis.
J.Biol.Chem., 298, 2022
|
|
3HGV
 
 | |
1V0D
 
 | | Crystal Structure of Caspase-activated DNase (CAD) | | Descriptor: | DNA FRAGMENTATION FACTOR 40 KDA SUBUNIT, LEAD (II) ION, MAGNESIUM ION, ... | | Authors: | Woo, E.-J, Kim, Y.-G, Kim, M.-S, Han, W.-D, Shin, S, Oh, B.-H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for Inactivation and Activation of Cad/Dff40 in the Apoptotic Pathway
Mol.Cell, 14, 2004
|
|
3JTL
 
 | |
4QU4
 
 | |
