1BSF
 
 | |
1BKP
 
 | |
1BSP
 
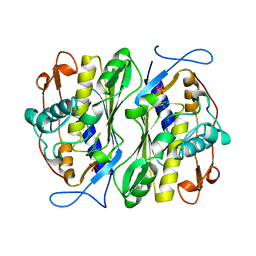 | | THERMOSTABLE THYMIDYLATE SYNTHASE A FROM BACILLUS SUBTILIS | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE A | | Authors: | Stout, T.J, Schellenberger, U, Santi, D.V, Stroud, R.M. | | Deposit date: | 1998-07-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of a unique thermal-stable thymidylate synthase from Bacillus subtilis.
Biochemistry, 37, 1998
|
|
9EQF
 
 | | Crystal structure of the L-arginine hydroxylase VioC MeHis316, bound to Fe(II), L-arginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Hardy, F.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Ferryl Reactivity in a Nonheme Iron Oxygenase Using an Expanded Genetic Code.
Acs Catalysis, 14, 2024
|
|
5K8H
 
 | |
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8H92
 
 | |
9J7N
 
 | | Cryo-EM structure of TauT | | Descriptor: | BETA-ALANINE, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J7M
 
 | | Cryo-EM structure of TauT | | Descriptor: | 2-AMINOETHANESULFONIC ACID, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J7O
 
 | | Cryo-EM structure of TauT | | Descriptor: | CHLORIDE ION, CHOLESTEROL, HEXADECANE, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9JQT
 
 | | Structure of interleukin receptor common gamma chain (IL2Rgamma/CD132) in complex with 2D4 | | Descriptor: | 2D4-Heavy Chain, 2D4-Light Chain, Anti-Fab nanobody, ... | | Authors: | Lu, Q.J, Yin, H.Q. | | Deposit date: | 2024-09-28 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | 2D4, a humanized monoclonal antibody targeting CD132, is a promising treatment for systemic lupus erythematosus.
Signal Transduct Target Ther, 9, 2024
|
|
7XCZ
 
 | |
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | Authors: | Liu, Z, Lui, S, Gao, Y. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | Authors: | Liu, Z, Liu, S, Gao, Y.Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDA
 
 | |
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
9J7D
 
 | | Arabidopsis high-affinity urea transport DUR3 in the inward-facing open conformation, dimeric state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, HEXADECANE, TETRADECANE, ... | | Authors: | An, W, Gao, Y, Zhang, X.C. | | Deposit date: | 2024-08-18 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of urea transport by Arabidopsis thaliana DUR3.
Nat Commun, 16, 2025
|
|
9J7C
 
 | | Arabidopsis high-affinity urea transport DUR3 in the urea-bound occluded conformation, dimeric state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, HEXADECANE, TETRADECANE, ... | | Authors: | An, W, Gao, Y, Zhang, X.C. | | Deposit date: | 2024-08-18 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of urea transport by Arabidopsis thaliana DUR3.
Nat Commun, 16, 2025
|
|
5YS1
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
5YS5
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant with seven copper ions | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-13 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
8JCN
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 58 | | Descriptor: | 1-[3-(diphenoxyphosphorylamino)phenyl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCK
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 32 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCM
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 55 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, HYDROSULFURIC ACID, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCO
 
 | |
