7DKD
 
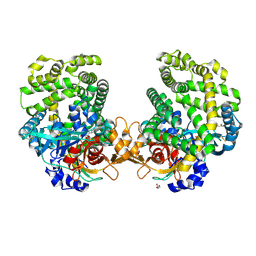 | | Stenotrophomonas maltophilia DPP7 in complex with Asn-Tyr | | Descriptor: | ASPARAGINE, Dipeptidyl-peptidase, GLYCEROL, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKC
 
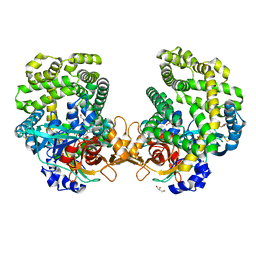 | | Stenotrophomonas maltophilia DPP7 in complex with Tyr-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, TYROSINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKE
 
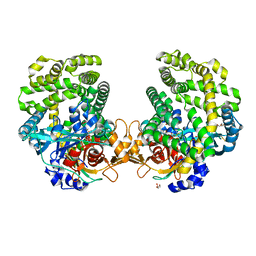 | | Stenotrophomonas maltophilia DPP7 in complex with Phe-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, PHENYLALANINE, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKB
 
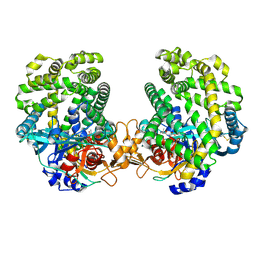 | | Stenotrophomonas maltophilia DPP7 in complex with Val-Tyr | | Descriptor: | Dipeptidyl-peptidase, TYROSINE, VALINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
3WON
 
 | | Crystal structure of the DAP BII dipeptide complex III | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOL
 
 | | Crystal structure of the DAP BII dipeptide complex I | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOO
 
 | | Crystal structure of the DAP BII hexapeptide complex I | | Descriptor: | Angiotensin II, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOK
 
 | | Crystal structure of the DAP BII (Space) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOJ
 
 | | Crystal structure of the DAP BII | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOP
 
 | | Crystal structure of the DAP BII hexapeptide complex II | | Descriptor: | Angiotensin IV, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOQ
 
 | | Crystal structure of the DAP BII hexapeptide complex III | | Descriptor: | Angiotensin IV, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
7DCZ
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-4H-1,3-thiazin-4-yl]-4- fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Koriyama, Y, Hori, A, Ito, H, Yonezawa, S, Baba, Y, Tanimoto, N, Ueno, T, Yamamoto, S, Yamamoto, T, Asada, N, Morimoto, K, Einaru, S, Sakai, K, Kanazu, T, Matsuda, A, Yamaguchi, Y, Oguma, T, Timmers, M, Tritsmans, L, Kusakabe, K.I, Kato, A, Sakaguchi, G. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Atabecestat (JNJ-54861911): A Thiazine-Based beta-Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor Advanced to the Phase 2b/3 EARLY Clinical Trial.
J.Med.Chem., 64, 2021
|
|
6CON
 
 | | Crystal structure of Mycobacterium tuberculosis IpdAB | | Descriptor: | CoA-transferase subunit alpha, CoA-transferase subunit beta | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CO6
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB | | Descriptor: | GLYCEROL, Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CO9
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6COJ
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB E105A COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3WOR
 
 | | Crystal structure of the DAP BII octapeptide complex | | Descriptor: | Angiotensin II, GLYCEROL, ZINC ION, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOM
 
 | | Crystal structure of the DAP BII dipeptide complex II | | Descriptor: | GLYCEROL, TYROSINE, VALINE, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WOI
 
 | | Crystal structure of the DAP BII (S657A) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WA6
 
 | |
2BL5
 
 | | Solution structure of the KH-QUA2 region of the Xenopus STAR-GSG Quaking protein. | | Descriptor: | MGC83862 PROTEIN | | Authors: | Maguire, M.L, Guler-Gane, G, Nietlispach, D, Raine, A.R.C, Zorn, A.M, Standart, N, Broadhurst, R.W. | | Deposit date: | 2005-03-01 | | Release date: | 2005-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Kh-Qua2 Region of the Xenopus Star/Gsg Quaking Protein
J.Mol.Biol., 348, 2005
|
|
3WA7
 
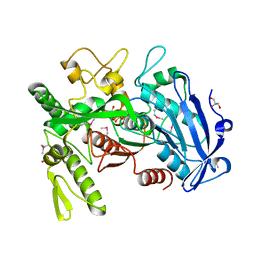 | | Crystal structure of selenomethionine-labeled tannase from Lactobacillus plantarum in the orthorhombic crystal | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Matoba, Y, Tanaka, N, Sugiyama, M. | | Deposit date: | 2013-04-27 | | Release date: | 2013-07-24 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic and mutational analyses of tannase from Lactobacillus plantarum.
Proteins, 81, 2013
|
|
2DFC
 
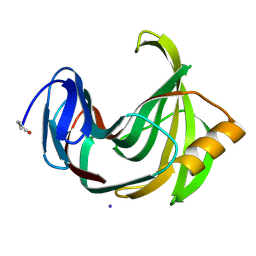 | | Xylanase II from Tricoderma reesei at 293K | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Harata, K, Akiba, T. | | Deposit date: | 2006-02-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DFB
 
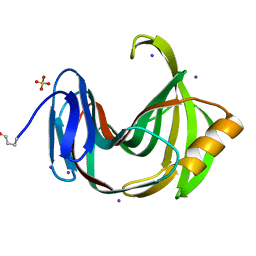 | | Xylanase II from Tricoderma reesei at 100K | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, SULFATE ION | | Authors: | Harata, K, Akiba, T. | | Deposit date: | 2006-02-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2M2Q
 
 | | Solution structure of MCh-1: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-1 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
