2CSU
 
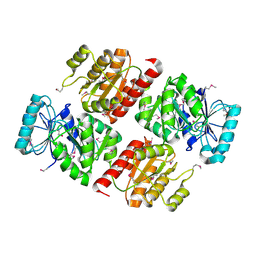 | |
2CU2
 
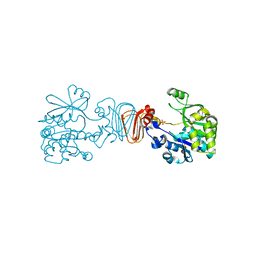 | |
2CUY
 
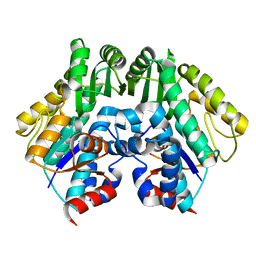 | | Crystal structure of malonyl CoA-acyl carrier protein transacylase from Thermus thermophilus HB8 | | Descriptor: | Malonyl CoA-[acyl carrier protein] transacylase | | Authors: | Misaki, S, Suzuki, K, Kunishima, N, Sugawara, M, Kuroishi, C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of malonyl CoA-acyl carrier protein transacylase from Thermus thermophilus HB8
TO BE PUBLISHED
|
|
2D1C
 
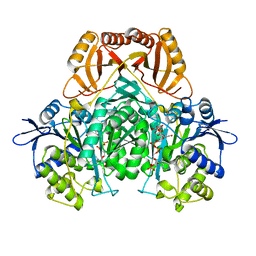 | |
2CU0
 
 | |
2CUK
 
 | |
2CY0
 
 | |
2DSG
 
 | | Crystal structure of Lys26 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2DJZ
 
 | |
2DV7
 
 | | Crystal structure of Lys187 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-28 | | Release date: | 2007-01-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2EEN
 
 | |
2E15
 
 | |
2EGL
 
 | | Crystal structure of Glu171 to Lys mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-01 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2EHL
 
 | |
2E66
 
 | | Crystal Structure Of CutA1 From Pyrococcus Horikoshii OT3, Mutation D60A | | Descriptor: | CHLORIDE ION, Divalent-cation tolerance protein cutA, SODIUM ION | | Authors: | Bagautdinov, B, Sawano, M, Bagautdinova, S, Yutani, K, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the hyper-thermostability of CutA1
To be Published
|
|
2EJA
 
 | |
2EKA
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L202M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Matsuura, Y, Kageyama, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L202M)
To be Published
|
|
2EV9
 
 | | Crystal Structure of Shikimate 5-Dehydrogenase (AroE) from Thermus Thermophilus HB8 in complex with NADP(H) and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Bagautdinov, B, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-31 | | Release date: | 2006-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Shikimate Dehydrogenase AroE from Thermus thermophilus HB8 and its Cofactor and Substrate Complexes: Insights into the Enzymatic Mechanism
J.Mol.Biol., 373, 2007
|
|
1HSR
 
 | | BINDING MODE OF BENZHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding mode of benzhydroxamic acid to Arthromyces ramosus peroxidase shown by X-ray crystallographic analysis of the complex at 1.6 A resolution.
FEBS Lett., 412, 1997
|
|
1CK6
 
 | | BINDING MODE OF SALICYLHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN (PEROXIDASE), ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1999-04-28 | | Release date: | 1999-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics.
Biochemistry, 38, 1999
|
|
1C8I
 
 | | BINDING MODE OF HYDROXYLAMINE TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | Authors: | Wariishi, H, Nonaka, D, Johjima, T, Nakamura, N, Naruta, Y, Kubo, K, Fukuyama, K. | | Deposit date: | 2000-05-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct binding of hydroxylamine to the heme iron of Arthromyces ramosus peroxidase. Substrate analogue that inhibits compound I formation in a competetive manner.
J.Biol.Chem., 275, 2000
|
|
1GZA
 
 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of iodide to Arthromyces ramosus peroxidase investigated with X-ray crystallographic analysis, 1H and 127I NMR spectroscopy, and steady-state kinetics.
J.Biol.Chem., 272, 1997
|
|
4RVC
 
 | |
5B35
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T, Suzuki, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B34
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with Iodine-labeled Detergent HAD13a Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2,4,6-tris(iodanyl)-5-(octanoylamino)benzene-1,3-dicarboxylic acid, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
