6B8U
 
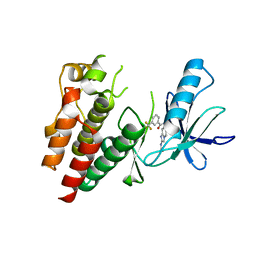 | | Crystals Structure of B-Raf kinase domain in complex with an Imidazopyridinyl benzamide inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Appleton, B.A, Murray, J, Shafer, C.M. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6KAF
 
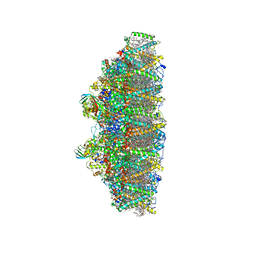 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4J99
 
 | |
4J96
 
 | |
8V52
 
 | |
9JLM
 
 | | Crystal structure of aldolase AtoB 1.9A | | Descriptor: | AtoB aldolase, CALCIUM ION | | Authors: | Ma, K, Fan, A, Lin, W. | | Deposit date: | 2024-09-19 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structural alignment based discovery and molecular basis of AtoB, catalyzing linear tetracyclic formation.
Chem Sci, 15, 2024
|
|
9IR5
 
 | |
9IR6
 
 | | Crystal structure of UDP-N-acetylmuramic Acid L-alanine ligase (MurC) from Roseburia faecis in complex with UNAM | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Wang, Y.X, Du, Y.H. | | Deposit date: | 2024-07-14 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Unusual MurC Ligase and Peptidoglycan Discovered in Lachnospiraceae Using a Fluorescent L-Amino Acid Based Selective Labeling Probe.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9J7N
 
 | | Cryo-EM structure of TauT | | Descriptor: | BETA-ALANINE, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J7M
 
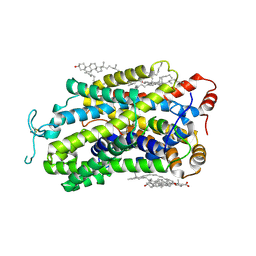 | | Cryo-EM structure of TauT | | Descriptor: | 2-AMINOETHANESULFONIC ACID, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J7O
 
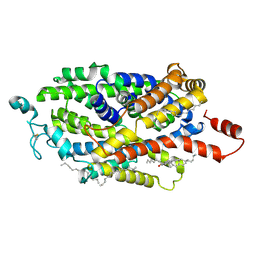 | | Cryo-EM structure of TauT | | Descriptor: | CHLORIDE ION, CHOLESTEROL, HEXADECANE, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J11
 
 | | Structure of mEos3.2 in the green fluorescent state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Green to red photoconvertible GFP-like protein EosFP | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9J0R
 
 | | Structure of pcStar in the green fluorescent state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Green to red photoconvertible GFP-like protein EosFP | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9J0Q
 
 | | Structure of mEos3.2 in the green fluorescent state | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7CH1
 
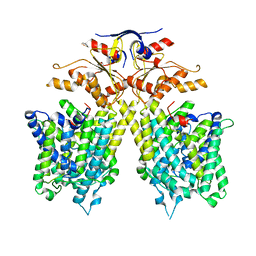 | | The overall structure of SLC26A9 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute carrier family 26 member 9 | | Authors: | Chi, X.M, Chen, Y, Li, X.R, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the gating mechanism of human SLC26A9 mediated by its C-terminal sequence.
Cell Discov, 6, 2020
|
|
8WMA
 
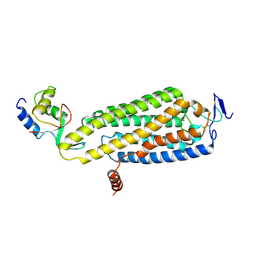 | | Fzd4/DEP complex (local refined) | | Descriptor: | Frizzled-4, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of Frizzled 4 in recognition of Dishevelled 2 unveils mechanism of WNT signaling activation.
Nat Commun, 15, 2024
|
|
8WM9
 
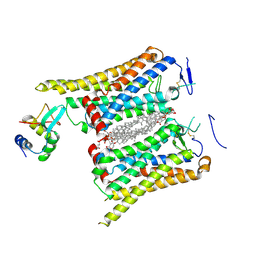 | | Fzd4/DEP complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Frizzled-4, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of Frizzled 4 in recognition of Dishevelled 2 unveils mechanism of WNT signaling activation.
Nat Commun, 15, 2024
|
|
7WVP
 
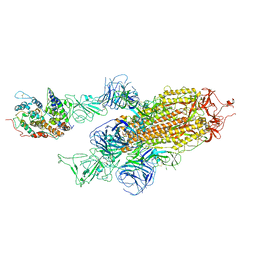 | |
7WVQ
 
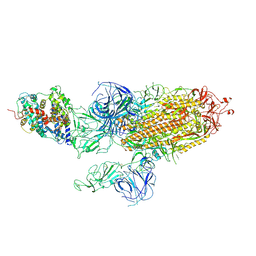 | |
8K9X
 
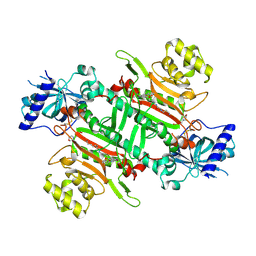 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 5 (ADKI5) | | Descriptor: | (2~{S})-2,6-bis(azanyl)-~{N}-[3-[2-[[4-[(2,5-dimethoxyphenyl)amino]-1,3,5-triazin-2-yl]amino]phenyl]sulfonylpropyl]hexanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lysine--tRNA ligase | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9V
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 3 (ADKI3) | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9S
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 1 (ADKI1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9W
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 4 (ADKI4) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lysine--tRNA ligase, ~{N}2-(2-methoxyphenyl)-~{N}4-(2-propan-2-ylsulfonylphenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
8K9U
 
 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 2 (ADKI2) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
7XM9
 
 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | Descriptor: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
