7JQ4
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI7 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ1
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7KPT
 
 | | Crystal structure of CtdE in complex with FAD and substrate 4 | | Descriptor: | (6aR,7aS,11S,13aS)-6,6,11-trimethyl-4-(3-methylbut-2-en-1-yl)-6,6a,7,8,9,10,11,14-octahydro-5H,13H-13a,7a-(epiminomethano)quinolizino[2,3-b]carbazol-16-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
7KPQ
 
 | | Crystal structure of CtdE in complex with FAD | | Descriptor: | FAD-dependent monooxygenase CtdE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
7JPZ
 
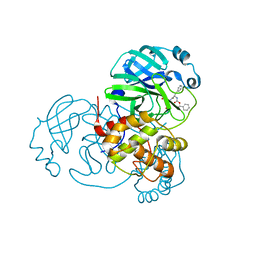 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI1 | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ3
 
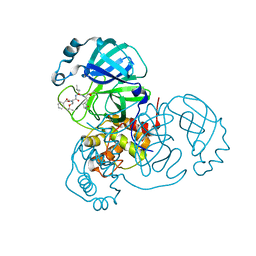 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI6 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ5
 
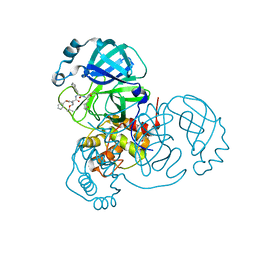 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
3MBD
 
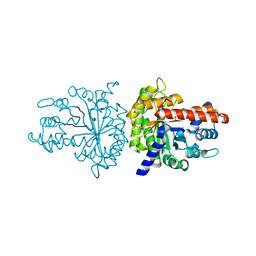 | |
3N58
 
 | |
3OCA
 
 | |
7LV3
 
 | | Crystal structure of human protein kinase G (PKG) R-C complex in inhibited state | | Descriptor: | 1,2-ETHANEDIOL, Isoform Beta of cGMP-dependent protein kinase 1, MANGANESE (II) ION, ... | | Authors: | Sharma, R, Lying, Q, Casteel, D, Kim, C. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | An auto-inhibited state of protein kinase G and implications for selective activation.
Elife, 11, 2022
|
|
7MBJ
 
 | |
4S2M
 
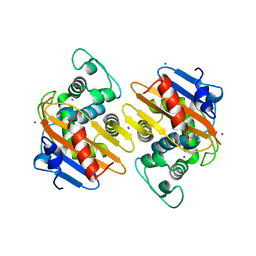 | | Crystal Structure of OXA-163 complexed with iodide in the active site | | Descriptor: | Beta-lactamase, IODIDE ION | | Authors: | Stojanoski, V, Hu, L, Palzkill, T.G, Prasad, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
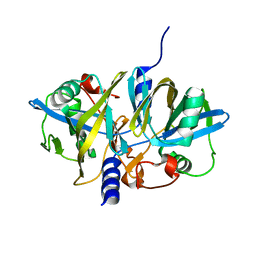
3NFW
 
 | |
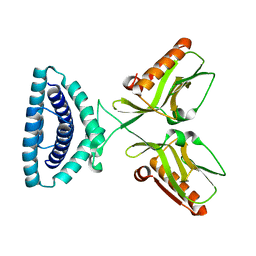
4OPA
 
 | |
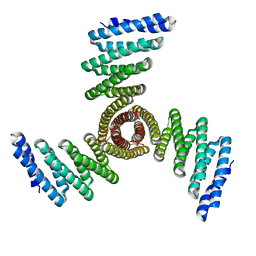
8FBO
 
 | |
8FBJ
 
 | |
4Y0V
 
 | |
8FBI
 
 | |
8FBK
 
 | |
8FBN
 
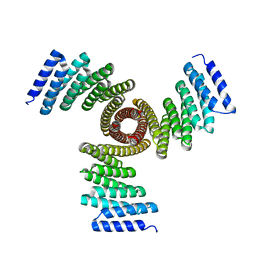 | |
8FBT
 
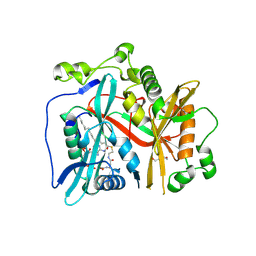 | | Crystal structure of Cryptosporidium parvum N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Staker, B.L, Fenwick, M.K, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of and Structural Insights into Hit Compounds Targeting N -Myristoyltransferase for Cryptosporidium Drug Development.
Acs Infect Dis., 9, 2023
|
|
8FBM
 
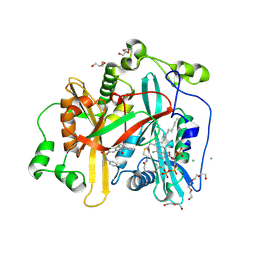 | |
8FBU
 
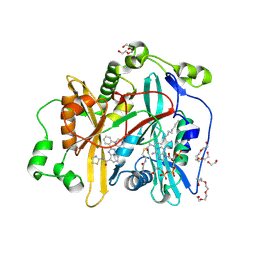 | | Crystal structure of Cryptosporidium parvum N-myristoyltransferase with bound myristoyl-CoA and Compound-2 | | Descriptor: | (2M)-2-[2-(piperazin-1-yl)phenyl]-N-(1,3-thiazol-2-yl)-1H-benzimidazole-4-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Staker, B.L, Fenwick, M.K, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of and Structural Insights into Hit Compounds Targeting N -Myristoyltransferase for Cryptosporidium Drug Development.
Acs Infect Dis., 9, 2023
|
|
3L3B
 
 | |
