3WBI
 
 | | Crystal structure analysis of eukaryotic translation initiation factor 5B structure I | | Descriptor: | Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WGL
 
 | | STAPHYLOCOCCUS AUREUS FTSZ T7 mutant substituted for GAN bound with GDP, DeltaT7GAN-GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Han, X, Matsui, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.066 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
3WKR
 
 | | Crystal structure of the SepCysS-SepCysE complex from Methanocaldococcus jannaschii | | Descriptor: | O-phospho-L-seryl-tRNA:Cys-tRNA synthase, Uncharacterized protein MJ1481 | | Authors: | Nakazawa, Y, Asano, N, Nakamura, A, Yao, M. | | Deposit date: | 2013-10-30 | | Release date: | 2014-07-16 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WT0
 
 | |
3W7A
 
 | | Crystal Structure of azoreductase AzrC fin complex with sulfone-modified azo dye Acid Red 88 | | Descriptor: | 4-[(E)-(2-hydroxynaphthalen-1-yl)diazenyl]naphthalene-1-sulfonic acid, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yu, J, Ogata, D, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WKH
 
 | | Crystal structure of cellobiose 2-epimerase in complex with epilactose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKS
 
 | | Crystal structure of the SepCysS-SepCysE N-terminal domain complex from | | Descriptor: | O-phospho-L-seryl-tRNA:Cys-tRNA synthase, Uncharacterized protein MJ1481 | | Authors: | Nakazawa, Y, Asano, N, Nakamura, A, Yao, M. | | Deposit date: | 2013-10-30 | | Release date: | 2014-07-30 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.029 Å) | | Cite: | Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3W77
 
 | | Crystal Structure of azoreductase AzrA | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Ogata, D, Yu, J, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WC1
 
 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with a G-1 deleted tRNA(His) | | Descriptor: | 75-mer tRNA, Likely histidyl tRNA-specific guanylyltransferase | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WC0
 
 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Likely histidyl tRNA-specific guanylyltransferase, MAGNESIUM ION | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WGN
 
 | | STAPHYLOCOCCUS AUREUS FTSZ bound with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Matsui, T, Mogi, N, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
3WGK
 
 | | STAPHYLOCOCCUS AUREUS FTSZ T7 mutant substituted for GAG, DeltaT7GAG-GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Han, X, Matsui, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
3WKI
 
 | | Crystal structure of cellobiose 2-epimerase in complex with cellobiitol | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WBZ
 
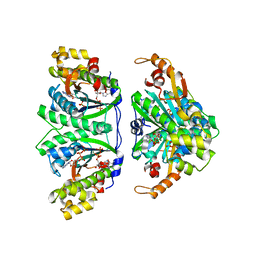 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Likely histidyl tRNA-specific guanylyltransferase, MAGNESIUM ION | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WBJ
 
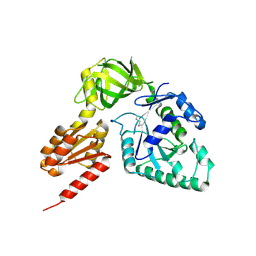 | | Crystal structure analysis of eukaryotic translation initiation factor 5B structure II | | Descriptor: | Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WC2
 
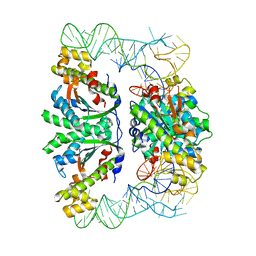 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with a tRNA(Phe)(GUG) | | Descriptor: | 76mer-tRNA, Likely histidyl tRNA-specific guanylyltransferase | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.641 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WKG
 
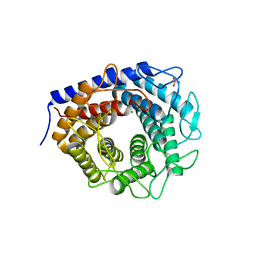 | | Crystal structure of cellobiose 2-epimerase in complex with glucosylmannose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKF
 
 | | Crystal structure of cellobiose 2-epimerase | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WY2
 
 | | Crystal structure of alpha-glucosidase in complex with glucose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WY9
 
 | | Crystal structure of a complex of the archaeal ribosomal stalk protein aP1 and the GDP-bound archaeal elongation factor aEF1alpha | | Descriptor: | 50S ribosomal protein L12, Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
3WY1
 
 | | Crystal structure of alpha-glucosidase | | Descriptor: | (3R,5R,7R)-octane-1,3,5,7-tetracarboxylic acid, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WYA
 
 | | Crystal structure of GDP-bound EF1alpha from Pyrococcus horikoshii | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
3WY3
 
 | | Crystal structure of alpha-glucosidase mutant D202N in complex with glucose and glycerol | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WY4
 
 | | Crystal structure of alpha-glucosidase mutant E271Q in complex with maltose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
1WE5
 
 | | Crystal Structure of Alpha-Xylosidase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative family 31 glucosidase yicI | | Authors: | Ose, T, Kitamura, M, Okuyama, M, Mori, H, Kimura, A, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2004-05-24 | | Release date: | 2005-02-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Alpha-Xylosidase from Escherichia coli
TO BE PUBLISHED
|
|
