6A6S
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans in complex with FSA, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
3A0N
 
 | | Crystal structure of D-glucuronic acid-bound alginate lyase vAL-1 from Chlorella virus | | Descriptor: | VAL-1, beta-D-glucopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-03-23 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
1Y3Q
 
 | | Structure of AlgQ1, alginate-binding protein | | Descriptor: | AlgQ1, CALCIUM ION | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1Y3P
 
 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate tetrasaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1Y3N
 
 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
1X1J
 
 | | Crystal Structure of Xanthan Lyase (N194A) with a Substrate. | | Descriptor: | (4AR,6R,7S,8R,8AR)-8-((5R,6R)-3-CARBOXY-TETRAHYDRO-4,5,6-TRIHYDROXY-2H-PYRAN-2-YLOXY)-HEXAHYDRO-6,7-DIHYDROXY-2-METHYLPYRANO[3,2-D][1,3]DIOXINE-2-CARBOXYLIC ACID), CALCIUM ION, xanthan lyase | | Authors: | Maruyama, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-04-04 | | Release date: | 2005-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism
J.Mol.Biol., 350, 2005
|
|
1X1H
 
 | |
1X1I
 
 | | Crystal Structure of Xanthan Lyase (N194A) Complexed with a Product | | Descriptor: | (4AR,6R,7S,8R,8AS)-HEXAHYDRO-6,7,8-TRIHYDROXY-2-METHYLPYRANO[3,2-D][1,3]DIOXINE-2-CARBOXYLIC ACID, xanthan lyase | | Authors: | Maruyama, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-04-04 | | Release date: | 2005-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism
J.Mol.Biol., 350, 2005
|
|
1VD5
 
 | | Crystal Structure of Unsaturated Glucuronyl Hydrolase, Responsible for the Degradation of Glycosaminoglycan, from Bacillus sp. GL1 at 1.8 A Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCINE, ... | | Authors: | Itoh, T, Akao, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-03-18 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Unsaturated Glucuronyl Hydrolase, Responsible for the Degradation of Glycosaminoglycan, from Bacillus sp. GL1 at 1.8 A Resolution
J.Biol.Chem., 279, 2004
|
|
1Y3I
 
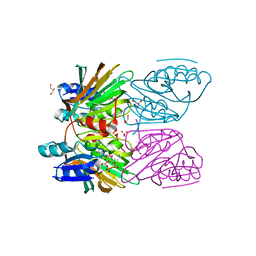 | | Crystal Structure of Mycobacterium tuberculosis NAD kinase-NAD complex | | Descriptor: | GLYCEROL, Inorganic polyphosphate/ATP-NAD kinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, Kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-25 | | Release date: | 2005-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
Biochem.Biophys.Res.Commun., 327, 2005
|
|
1Y3H
 
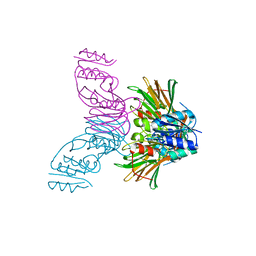 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD kinase from Mycobacterium tuberculosis | | Descriptor: | Inorganic polyphosphate/ATP-NAD kinase | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
BIOCHEM.BIOPHYS.RES.COMMUN., 327, 2005
|
|
2A4V
 
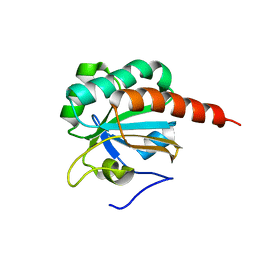 | | Crystal Structure of a truncated mutant of yeast nuclear thiol peroxidase | | Descriptor: | Peroxiredoxin DOT5 | | Authors: | Choi, J, Choi, S, Chon, J.-K, Choi, J, Cha, M.-K, Kim, I.-H, Shin, W. | | Deposit date: | 2005-06-29 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C107S/C112S mutant of yeast nuclear 2-Cys peroxiredoxin
Proteins, 61, 2005
|
|
2Z42
 
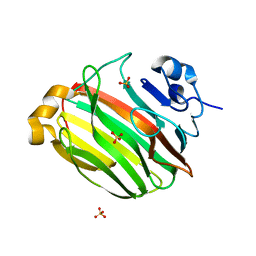 | | Crystal Structure of Family 7 Alginate Lyase A1-II' from Sphingomonas sp. A1 | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2007-06-12 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Recognition by Family 7 Alginagte Lyase from Sphingomonas sp. A1
To be published
|
|
2ZAB
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Cmplex with Product (GGG) | | Descriptor: | Alginate lyase, GLYCEROL, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZAC
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Complex with Product (MMG) | | Descriptor: | Alginate lyase, GLYCEROL, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZUY
 
 | | Crystal structure of exotype rhamnogalacturonan lyase YesX | | Descriptor: | CALCIUM ION, YesX protein | | Authors: | Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases
J.Biol.Chem., 284, 2009
|
|
2Z8R
 
 | | Crystal structure of rhamnogalacturonan lyase YesW at 1.40 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
2ZAA
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' H191N/Y284F in Complex with Substrate (GGMG) | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase, GLYCEROL | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2Z8S
 
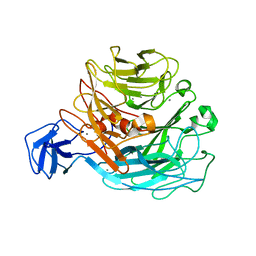 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with digalacturonic acid | | Descriptor: | CALCIUM ION, YesW protein, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
3A09
 
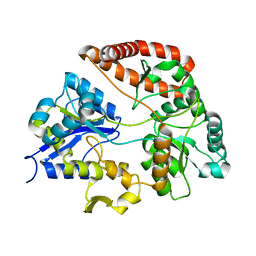 | |
2ZA9
 
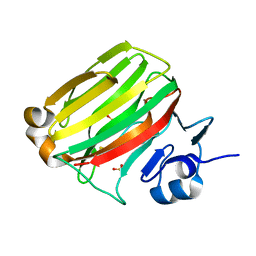 | | Crystal Structure of Alginate lyase A1-II' N141C/N199C | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZUX
 
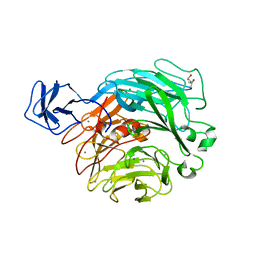 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein, ... | | Authors: | Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases
J.Biol.Chem., 284, 2009
|
|
3AFM
 
 | | Crystal structure of aldose reductase A1-R responsible for alginate metabolism | | Descriptor: | Carbonyl reductase | | Authors: | Takase, R, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-03-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1
Biochim.Biophys.Acta, 1804, 2010
|
|
3AMI
 
 | | The crystal structure of the M16B metallopeptidase subunit from Sphingomonas sp. A1 | | Descriptor: | zinc peptidase | | Authors: | Maruyama, Y, Chuma, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-08-20 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Heterosubunit composition and crystal structures of a novel bacterial M16B metallopeptidase
J.Mol.Biol., 407, 2011
|
|
3AMJ
 
 | | The crystal structure of the heterodimer of M16B peptidase from Sphingomonas sp. A1 | | Descriptor: | ZINC ION, zinc peptidase active subunit, zinc peptidase inactive subunit | | Authors: | Maruyama, Y, Chuma, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-08-20 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Heterosubunit composition and crystal structures of a novel bacterial M16B metallopeptidase
J.Mol.Biol., 407, 2011
|
|
