5M67
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenine and 2'-deoxyadenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, ACETATE ION, ADENINE, ... | | Authors: | Manszewski, T, Jaskolski, M. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
4JHG
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with trans-zeatin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, MALONATE ION, MtN13 protein, ... | | Authors: | Ruszkowski, M, Tusnio, K, Ciesielska, A, Brzezinski, K, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5EBI
 
 | | Crystal structure of a DNA-RNA chimera in complex with Ba2+ ions: a case of unusual multi-domain twinning | | Descriptor: | BARIUM ION, DNA/RNA (5'-D(*C)-R(P*G)-D(P*C)-R(P*G)-D(P*C)-R(P*G)-3') | | Authors: | Gilski, M, Drozdzal, P, Kierzek, R, Jaskolski, M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic resolution structure of a chimeric DNA-RNA Z-type duplex in complex with Ba(2+) ions: a case of complicated multi-domain twinning.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1NAR
 
 | |
1PZC
 
 | | APO-PSEUDOAZURIN (METAL FREE PROTEIN) | | Descriptor: | PSEUDOAZURIN | | Authors: | Petratos, K. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of apo-pseudoazurin from Alcaligenes faecalis S-6.
Febs Lett., 368, 1995
|
|
2OGR
 
 | |
2OJK
 
 | | Crystal Structure of Green Fluorescent Protein from Zoanthus sp at 2.2 A Resolution | | Descriptor: | GFP-like fluorescent chromoprotein FP506 | | Authors: | Pletneva, N.V, Pletnev, S.V, Tikhonova, T.V, Pletnev, V.Z. | | Deposit date: | 2007-01-12 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3GB3
 
 | |
3GL4
 
 | |
1H4G
 
 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | SULFATE ION, XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalysis and Specificity in Enzymatic Glycoside Hydrolysis: A 2,5B Conformation for the Glycosyl-Enzyme Intermediate Revealed by the Structure of the Bacillus Agaradhaerens Family 11 Xylanase.
Chem.Biol., 6, 1999
|
|
1H4H
 
 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis and Specificity in Enzymatic Glycoside Hydrolysis: A 2,5B Conformation for the Glycosyl-Enzyme Intermediate Revealed by the Structure of the Bacillus Agaradhaerens Family 11 Xylanase.
Chem.Biol., 6, 1999
|
|
2OXI
 
 | |
2PXW
 
 | | Crystal Structure of N66D Mutant of Green Fluorescent Protein from Zoanthus sp. at 2.4 A Resolution (Transition State) | | Descriptor: | GFP-like fluorescent chromoprotein FP506 | | Authors: | Pletnev, S.V, Pletneva, N.V, Tikhonova, T.V, Pletnev, V.Z. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2PXS
 
 | | Crystal Structure of N66D Mutant of Green Fluorescent Protein from Zoanthus sp. at 2.2 A Resolution (Mature State) | | Descriptor: | GFP-like fluorescent chromoprotein FP506 | | Authors: | Pletnev, S.V, Pletneva, N.V, Tikhonova, T.V, Pletnev, V.Z. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6DWF
 
 | |
6DWH
 
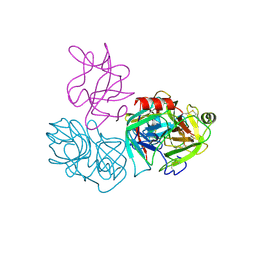 | | Crystal structure of complex of BBKI and Bovine Trypsin | | Descriptor: | CHLORIDE ION, Cationic trypsin, Kunitz-type inihibitor, ... | | Authors: | Li, M, Wlodawer, A. | | Deposit date: | 2018-06-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the complex of a kallikrein inhibitor from Bauhinia bauhinioides with trypsin and modeling of kallikrein complexes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6DWU
 
 | |
4Q7T
 
 | |
4Q7U
 
 | |
3U8C
 
 | |
5KY6
 
 | |
3U8A
 
 | |
1ERR
 
 | |
2B96
 
 | | Third Calcium ion found in an inhibitor bound phospholipase A2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-METHOXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Yamane, T, Tsai, M.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Third Calcium ion found in an inhibitor bound phospholipase A2
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1ERE
 
 | |
