1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MP4
 
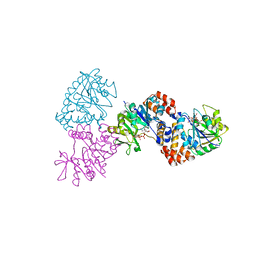 | | W224H VARIANT OF S. ENTERICA RmlA | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, W224H Variant of S. Enterica RmlA Bound to UDP-Glucose | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MHK
 
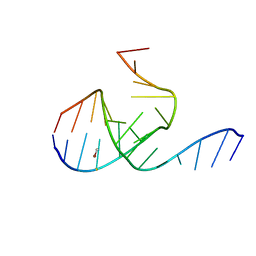 | | Crystal Structure Analysis of a 26mer RNA molecule, representing a new RNA motif, the hook-turn | | Descriptor: | BROMIDE ION, RNA 12-mer BCh12, RNA 14-mer BCh12 | | Authors: | Szep, S, Wang, J, Moore, P.B. | | Deposit date: | 2002-08-20 | | Release date: | 2002-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a 26-nucleotide RNA containing a hook-turn
RNA, 9, 2003
|
|
1MP3
 
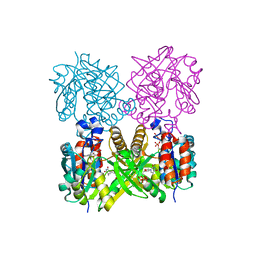 | | L89T VARIANT OF S. ENTERICA RmlA | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.T, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1R8B
 
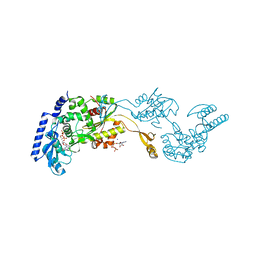 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1NIZ
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
1NJ2
 
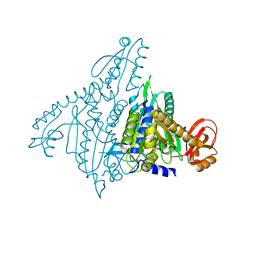 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus | | Descriptor: | MAGNESIUM ION, Proline-tRNA Synthetase, ZINC ION | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ5
 
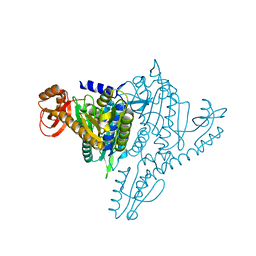 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to proline sulfamoyl adenylate | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ1
 
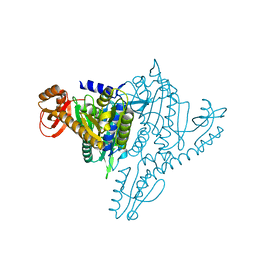 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to cysteine sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NJ8
 
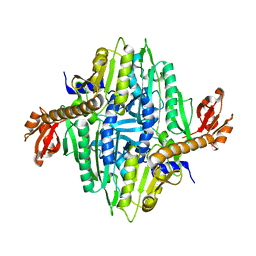 | | Crystal Structure of Prolyl-tRNA Synthetase from Methanocaldococcus janaschii | | Descriptor: | Proline-tRNA Synthetase | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N6U
 
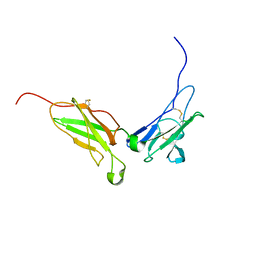 | | NMR structure of the interferon-binding ectodomain of the human interferon receptor | | Descriptor: | Interferon-alpha/beta receptor beta chain | | Authors: | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | Deposit date: | 2002-11-12 | | Release date: | 2003-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
1NGM
 
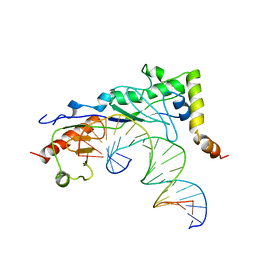 | | Crystal structure of a yeast Brf1-TBP-DNA ternary complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*AP*AP*AP*AP*AP*AP*AP*TP*GP*TP*TP*TP*TP*TP*T)-3', Transcription factor IIIB BRF1 subunit, ... | | Authors: | Juo, Z.S, Kassavetis, G.A, Wang, J, Geiduschek, E.P, Sigler, P.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a transcription factor IIIB core interface ternary complex
Nature, 422, 2003
|
|
1NJ0
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
1CJY
 
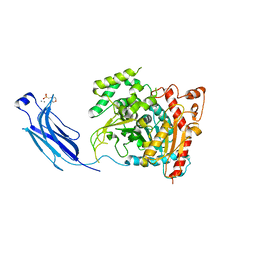 | | HUMAN CYTOSOLIC PHOSPHOLIPASE A2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, PROTEIN (CYTOSOLIC PHOSPHOLIPASE A2) | | Authors: | Dessen, A, Tang, J, Schmidt, H, Stahl, M, Clark, J.D, Seehra, J, Somers, W.S. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
2BTX
 
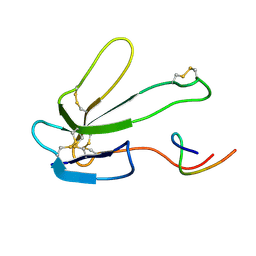 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF ALPHA-BUNGAROTOXIN WITH A LIBRARY DERIVED PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ALPHA-BUNGAROTOXIN, LIBRARY DERIVED PEPTIDE | | Authors: | Scherf, T, Balass, M, Fuchs, S, Katchalski-Katzir, E, Anglister, J. | | Deposit date: | 1998-08-23 | | Release date: | 1999-01-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the complex of alpha-bungarotoxin with a library-derived peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1L1F
 
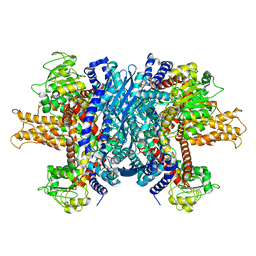 | | Structure of human glutamate dehydrogenase-apo form | | Descriptor: | Glutamate Dehydrogenase 1 | | Authors: | Smith, T.J, Schmidt, T, Fang, J, Wu, J, Siuzdak, G, Stanley, C.A. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of apo human glutamate dehydrogenase details subunit communication and allostery.
J.Mol.Biol., 318, 2002
|
|
1LJZ
 
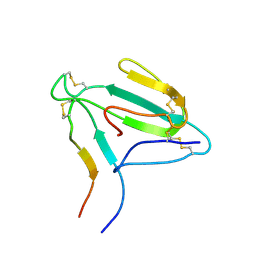 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Eisenstein, M, Chill, J.H, Anglister, J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1ZRR
 
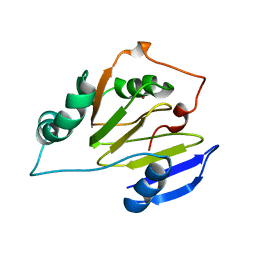 | | Residual Dipolar Coupling Refinement of Acireductone Dioxygenase from Klebsiella | | Descriptor: | E-2/E-2' protein, NICKEL (II) ION | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Ju, T, Hoefler, C, Liang, J. | | Deposit date: | 2005-05-19 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A refined model for the structure of acireductone dioxygenase from Klebsiella ATCC 8724 incorporating residual dipolar couplings
J.Biomol.NMR, 34, 2006
|
|
1L4W
 
 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Rodriguez, E, Eisenstein, M, Anglister, J. | | Deposit date: | 2002-03-06 | | Release date: | 2002-07-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1NJ6
 
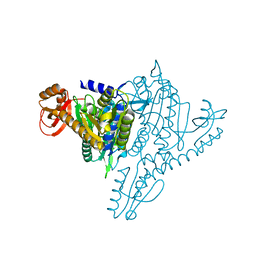 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to alanine sulfamoyl adenylate | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NSO
 
 | | Folded monomer of protease from Mason-Pfizer monkey virus | | Descriptor: | Protease 13 kDa | | Authors: | Veverka, V, Bauerova, H, Zabransky, A, Lang, J, Ruml, T, Pichova, I, Hrabal, R. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a monomeric form of a retroviral protease
J.MOL.BIOL., 333, 2003
|
|
1M1C
 
 | | Structure of the L-A virus | | Descriptor: | Major coat protein | | Authors: | Naitow, H, Tang, J, Canady, M, Wickner, R.B, Johnson, J.E. | | Deposit date: | 2002-06-18 | | Release date: | 2002-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism.
Nat.Struct.Biol., 9, 2002
|
|
1MIV
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme | | Descriptor: | MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1BRM
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 1998-08-24 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis.
J.Mol.Biol., 289, 1999
|
|
