4RWU
 
 | | J-domain of Sis1 protein, Hsp40 co-chaperone from Saccharomyces cerevisiae | | Descriptor: | Protein SIS1 | | Authors: | Osipiuk, J, Zhou, M, Gu, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-05 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Roles of intramolecular and intermolecular interactions in functional regulation of the Hsp70 J-protein co-chaperone Sis1.
J.Mol.Biol., 427, 2015
|
|
7TAI
 
 | | Structure of STEAP2 in complex with ligands | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, L, Chen, K.H, Zhou, M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-01-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of stepwise electron transfer in six-transmembrane epithelial antigen of the prostate (STEAP) 1 and 2.
Elife, 12, 2023
|
|
4ZO4
 
 | | Dephospho-CoA kinase from Campylobacter jejuni. | | Descriptor: | BETA-MERCAPTOETHANOL, Dephospho-CoA kinase | | Authors: | Osipiuk, J, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-06 | | Release date: | 2015-05-13 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Dephospho-CoA kinase from Campylobacter jejuni.
to be published
|
|
8ETB
 
 | | the crystal structure of a rationally designed zinc sensor based on maltose binding protein - Zn binding conformation | | Descriptor: | ACETATE ION, ZINC ION, Zinc Sensor protein | | Authors: | Zhao, Z, Zhou, M, Zemerov, S.D, Marmorstein, R, Dmochowski, I.J. | | Deposit date: | 2022-10-16 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
5INT
 
 | | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC | | Descriptor: | Phosphopantothenate--cysteine ligase | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC
To Be Published
|
|
8F23
 
 | | The crystal structure of a rationally designed zinc sensor based on maltose binding protein - Apo conformation | | Descriptor: | Zinc Sensor protein | | Authors: | Zhao, Z, Zhou, M, Zemerov, S.d, Marmorstein, R, Dmochowski, I.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
5JG7
 
 | | Crystal structure of putative periplasmic binding protein from Salmonella typhimurium LT2 | | Descriptor: | Fur regulated Salmonella iron transporter, GLYCEROL | | Authors: | Chang, C, Zhou, M, Shatsman, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative periplasmic binding protein from Salmonella typhimurium LT2
To Be Published
|
|
5IZN
 
 | | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6 | | Descriptor: | 50S ribosomal protein L25, PHOSPHATE ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-25 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6
To Be Published
|
|
5JCV
 
 | | Sortase B from Listeria monocytogenes. | | Descriptor: | CHLORIDE ION, Lmo2181 protein, SULFATE ION | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Sortase B from Listeria monocytogenes.
to be published
|
|
5CJJ
 
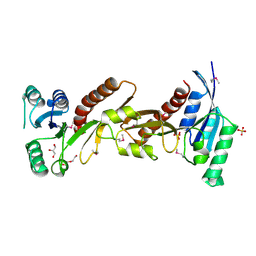 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-14 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To Be Published
|
|
4M0G
 
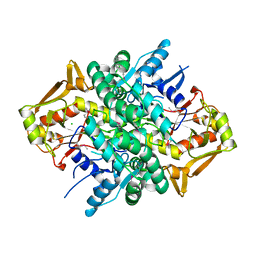 | | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor. | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Tan, K, Zhou, M, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor.
To be Published
|
|
7LZG
 
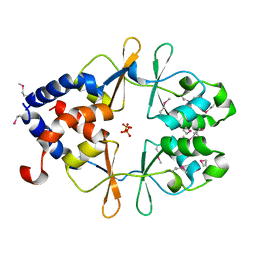 | |
5EUF
 
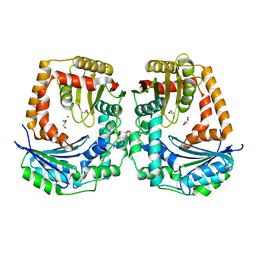 | | The crystal structure of a protease from Helicobacter pylori | | Descriptor: | GLYCEROL, Protease, ZINC ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a protease from Helicobacter pylori
To Be Published
|
|
5F1Q
 
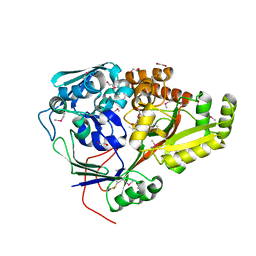 | | Crystal Structure of Periplasmic Dipeptide Transport Protein from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-30 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Crystal Structure of Periplasmic Dipeptide Transport Protein from Yersinia pestis
To Be Published
|
|
4NMU
 
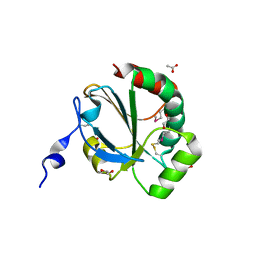 | | Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor'
To be Published
|
|
5EWQ
 
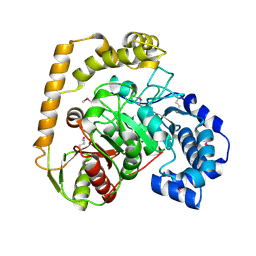 | | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames | | Descriptor: | ACETATE ION, Amidase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames
To Be Published
|
|
5EV7
 
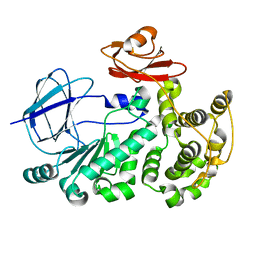 | | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames | | Descriptor: | Conserved domain protein | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames.
To Be Published
|
|
4NPB
 
 | | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92 | | Descriptor: | PHOSPHATE ION, Protein disulfide isomerase II, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92
To be Published
|
|
4NMW
 
 | | Crystal Structure of Carboxylesterase BioH from Salmonella enterica | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pimelyl-[acyl-carrier protein] methyl ester esterase | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Crystal Structure of Carboxylesterase BioH from Salmonella enterica
To be Published
|
|
4MA0
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP
To be Published
|
|
4M9U
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4MA5
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP. | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP.
To be Published
|
|
4MAM
 
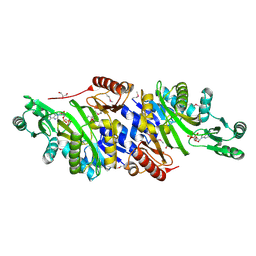 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-16 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP
To be Published
|
|
4NMY
 
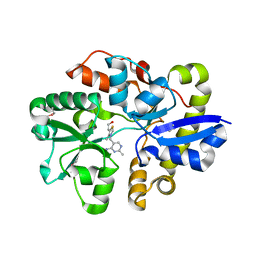 | | Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ABC-type transport system, extracellular solute-binding protein | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal Structure of the Thiamin-bound form of Substrate-binding Protein of ABC Transporter from Clostridium difficile
To be Published
|
|
4NP6
 
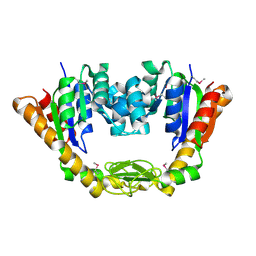 | | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor | | Descriptor: | Adenylate kinase | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of Adenylate Kinase from Vibrio cholerae O1 biovar eltor
To be Published
|
|
