7PQN
 
 | | Catalytic fragment of MASP-2 in complex with ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 2 A chain, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.400015 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
5E78
 
 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
7P4N
 
 | |
5DLB
 
 | | Crystal structure of chaperone EspG3 of ESX-3 type VII secretion system from Mycobacterium marinum M | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Chan, S, Arbing, M.A, Kim, J, Kahng, S, Sawaya, M.R, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Variability of EspG Chaperones from Mycobacterial ESX-1, ESX-3, and ESX-5 Type VII Secretion Systems.
J. Mol. Biol., 431, 2019
|
|
4RCL
 
 | |
2XZS
 
 | | Death associated protein kinase 1 residues 1-312 | | Descriptor: | DEATH ASSOCIATED KINASE 1, MAGNESIUM ION | | Authors: | Yumerefendi, H, Mas, P.J, Dordevic, N, McCarthy, A.A, Hart, D.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Death-Associated Protein Kinase Activity is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure, 24, 2016
|
|
4L4W
 
 | |
4KXR
 
 | |
2O2W
 
 | |
4BEG
 
 | |
2BZR
 
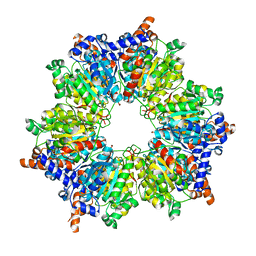 | |
6SUI
 
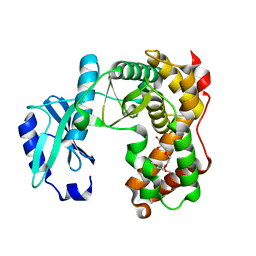 | | AMICOUMACIN KINASE AMIN | | Descriptor: | PENTAETHYLENE GLYCOL, Phosphotransferase enzyme family protein | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SUN
 
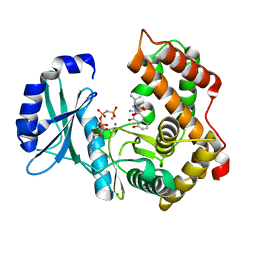 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, Ca2+ and Ami | | Descriptor: | APH domain-containing protein, amicoumacin kinase, Amicoumacin A, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6TGN
 
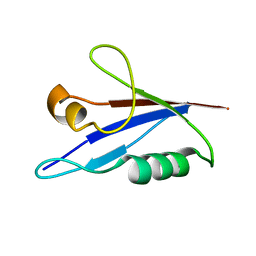 | |
6TH3
 
 | |
6SV5
 
 | | Amicoumacin kinase AmiN in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphotransferase enzyme family protein, amicoumacin kinase | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6TGY
 
 | |
6SUM
 
 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, MG2+ and Ami | | Descriptor: | ACETATE ION, AMICOUMACIN KINASE, Amicoumacin A, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6TGP
 
 | |
6SUL
 
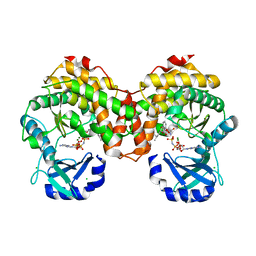 | | Amicoumacin kinase AmiN in complex with AMP-PNP, Mg2+ and Ami | | Descriptor: | Amicoumacin A, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6TGS
 
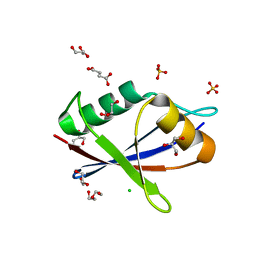 | | AtNBR1-PB1 domain | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Jakobi, A.J, Sachse, C. | | Deposit date: | 2019-11-17 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake.
Nat Commun, 11, 2020
|
|
2FGO
 
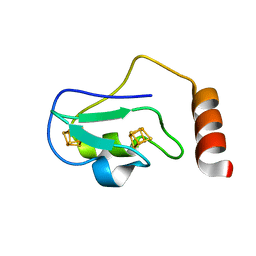 | | Structure of the 2[4FE-4S] ferredoxin from Pseudomonas aeruginosa | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER | | Authors: | Giastas, P, Pinotsis, N, Mavridis, I.M. | | Deposit date: | 2005-12-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structure of the 2[4Fe-4S] ferredoxin from Pseudomonas aeruginosa at 1.32-A resolution: comparison with other high-resolution structures of ferredoxins and contributing structural features to reduction potential values.
J.Biol.Inorg.Chem., 11, 2006
|
|
1AWO
 
 | |
2A38
 
 | | Crystal structure of the N-Terminus of titin | | Descriptor: | CADMIUM ION, Titin | | Authors: | Marino, M, Muhle-Goll, C, Svergun, D, Demirel, M, Mayans, O. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ig doublet Z1Z2: a model system for the hybrid analysis of conformational dynamics in Ig tandems from titin
Structure, 14, 2006
|
|
1AEY
 
 | |
