8Z3J
 
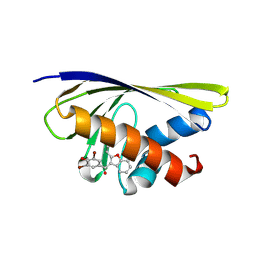 | | Benzbromarone interferes with the interaction between Hsp90 and Aha1 by interacting with both of them | | Descriptor: | Activator of 90 kDa heat shock protein ATPase homolog 1, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Shi, L, Zhong, Y, Tang, J, Xu, Z, Zhang, N. | | Deposit date: | 2024-04-15 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Benzbromarone interferes with the interaction between Hsp90 and Aha1 by interacting with both of them
To Be Published
|
|
8Z3H
 
 | |
1O19
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-15 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
7VJT
 
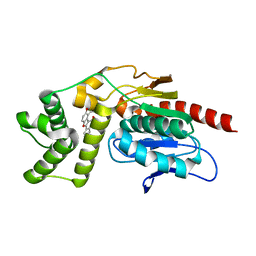 | | Crystal Structure of Mtb Pks13-TE in complex with inhibitor coumestan derivative 8 | | Descriptor: | 3,8-bis(oxidanyl)-7-(piperidin-1-ylmethyl)-[1]benzofuro[3,2-c]chromen-6-one, Polyketide synthase Pks13 (Termination polyketide synthase) | | Authors: | Zhang, W, Wang, S.S, Yu, L.F. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of Coumestan Derivatives as Polyketide Synthase 13-Thioesterase(Pks13-TE) Inhibitors with Improved hERG Profiles for Mycobacterium tuberculosis Treatment.
J.Med.Chem., 65, 2022
|
|
1O1A
 
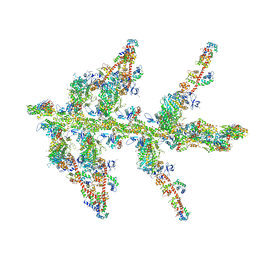 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-18 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O18
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-04 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1F
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-15 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1B
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-15 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1C
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-18 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1G
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle.
J.Struct.Biol., 138, 2002
|
|
1M8Q
 
 | | Molecular Models of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle | | Descriptor: | Skeletal muscle Actin, Skeletal muscle Myosin II, Skeletal muscle Myosin II Essential Light Chain, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-10 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle.
J.Struct.Biol., 138, 2002
|
|
1O1E
 
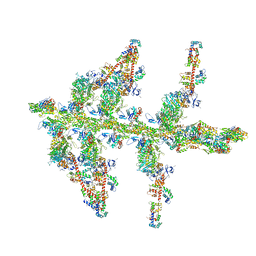 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O1D
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-18 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1MVW
 
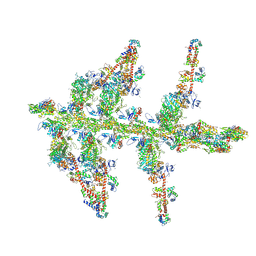 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | RABBIT SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL; LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-09-26 | | Release date: | 2002-11-20 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.STRUCT.BIOL., 138, 2002
|
|
7FAH
 
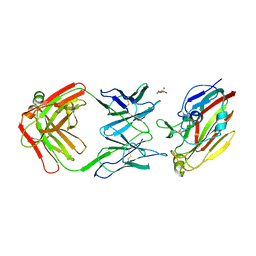 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
7DBF
 
 | |
7DC9
 
 | |
7DCA
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase bound by xanthosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-10-23 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The C-terminal loop of Arabidopsis thaliana guanosine deaminase is essential to catalysis.
Chem.Commun.(Camb.), 57, 2021
|
|
7DCB
 
 | |
7DCW
 
 | |
7DQN
 
 | |
7DH1
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N2-Methylguanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-12 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DM5
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with guanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DM6
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase complexed with crotonoside | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1H-purin-2-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
7DGC
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 2'-O-Methylguanosine | | Descriptor: | 9-[(2R,3R,4R)-5-(hydroxymethyl)-3-methoxy-4-oxidanyl-oxolan-2-yl]-3H-purine-2,6-dione, Guanosine deaminase, SODIUM ION, ... | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
