4IGL
 
 | | Structure of the RHS-repeat containing BC component of the secreted ABC toxin complex from Yersinia entomophaga | | Descriptor: | GLYCEROL, POTASSIUM ION, YenB, ... | | Authors: | Busby, J.N, Panjikar, S, Landsberg, M.J, Hurst, M.R.H, Lott, J.S. | | Deposit date: | 2012-12-17 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device
Nature, 501, 2013
|
|
4MGH
 
 | | Importance of Hydrophobic Cavities in Allosteric Regulation of Formylglycinamide Synthetase: Insight from Xenon Trapping and Statistical Coupling Analysis | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tanwar, A.S, Goyal, V.D, Choudhary, D, Panjikar, S, Anand, R. | | Deposit date: | 2013-08-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Importance of hydrophobic cavities in allosteric regulation of formylglycinamide synthetase: insight from xenon trapping and statistical coupling analysis
Plos One, 8, 2013
|
|
2JCG
 
 | | Apo form of the catabolite control protein A (ccpA) from bacillus megaterium, with the DNA binding domain | | Descriptor: | CALCIUM ION, GLUCOSE-RESISTANCE AMYLASE REGULATOR | | Authors: | Singh, R.K, Panjikar, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Apo Form of the Catabolite Control Protein a (Ccpa) from Bacillus Megaterium with a DNA-Binding Domain.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2JF7
 
 | | Structure of Strictosidine Glucosidase | | Descriptor: | STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
2JKG
 
 | | Plasmodium falciparum profilin | | Descriptor: | MAGNESIUM ION, OCTAPROLINE PEPTIDE, PROFILIN | | Authors: | Kursula, I, Kursula, P, Ganter, M, Panjikar, S, Matuschewski, K, Schueler, H. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis for Parasite-Specific Functions of the Divergent Profilin of Plasmodium Falciparum.
Structure, 16, 2008
|
|
2JKF
 
 | | Plasmodium falciparum profilin | | Descriptor: | PROFILIN | | Authors: | Kursula, I, Kursula, P, Ganter, M, Panjikar, S, Matuschewski, K, Schueler, H. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Parasite-Specific Functions of the Divergent Profilin of Plasmodium Falciparum
Structure, 16, 2008
|
|
7R0J
 
 | | Structure of the V2 receptor Cter-arrestin2-ScFv30 complex | | Descriptor: | Arrestin2, ScFv30, V2R Cter | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
7R0C
 
 | | Structure of the AVP-V2R-arrestin2-ScFv30 complex | | Descriptor: | AVP, Arrestin2, ScFv30, ... | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
3P5C
 
 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | Descriptor: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
3P86
 
 | |
3PPZ
 
 | |
1HL3
 
 | | CtBP/BARS in ternary complex with NAD(H) and PIDLSKK peptide | | Descriptor: | C-TERMINAL BINDING PROTEIN 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PRO-ILE-ASP-LEU-SER-LYS-LYS PEPTIDE | | Authors: | Nardini, M, Spano, S, Cericola, C, Pesce, A, Massaro, A, Millo, E, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2003-03-13 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ctbp/Bars: A Dual-Function Protein Involved in Transcription Co-Repression and Golgi Membrane Fission
Embo J., 22, 2003
|
|
7BES
 
 | | CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) in complex with UDP and UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Bous, J, Trapani, S, Walter, P, Bron, P, Munier-Lehmann, H. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
3OC5
 
 | |
3OC8
 
 | | Crystal Structure of the C-terminal Domain of the Vibrio cholerae soluble colonization factor TcpF | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Craig, L, Kolappan, S, Yuen, A.S.W. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Vibrio cholerae Colonization Factor TcpF and Identification of a Functional Immunogenic Site.
J.Mol.Biol., 409, 2011
|
|
3P5B
 
 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | Descriptor: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-10-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ILX
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Singh, A, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution
To Be Published
|
|
2ALG
 
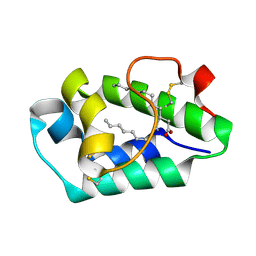 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
3E4P
 
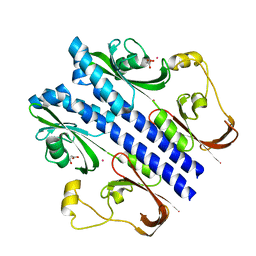 | | Crystal structure of malonate occupied DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MALONIC ACID, STRONTIUM ION | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
3E4O
 
 | | Crystal structure of succinate bound state DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MAGNESIUM ION, SUCCINIC ACID | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
3E4Q
 
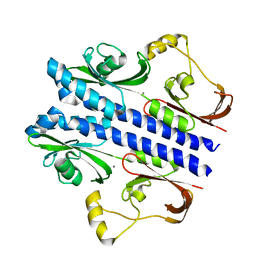 | | Crystal structure of apo DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, CALCIUM ION | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
3GZF
 
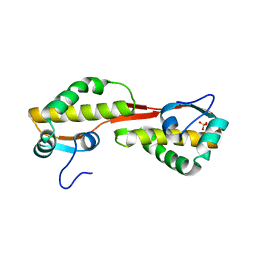 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6HJM
 
 | |
6AC3
 
 | |
