4LDM
 
 | |
3VH3
 
 | | Crystal structure of Atg7CTD-Atg8 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Noda, N.N, Satoo, K, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
3VH4
 
 | | Crystal structure of Atg7CTD-Atg8-MgATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autophagy-related protein 8, MAGNESIUM ION, ... | | Authors: | Noda, N.N, Satoo, K, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
4LDI
 
 | |
4LDB
 
 | |
4LD8
 
 | |
7W36
 
 | |
7WJT
 
 | |
7YDO
 
 | | Crystal structure of Atg44 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Uncharacterized protein C26A3.14c | | Authors: | Maruyama, T, Noda, N.N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mitochondrial intermembrane space protein mitofissin drives mitochondrial fission required for mitophagy.
Mol.Cell, 83, 2023
|
|
6JSA
 
 | |
6JS9
 
 | |
6JSC
 
 | |
6JSD
 
 | |
6JSB
 
 | |
1V3X
 
 | | Factor Xa in complex with the inhibitor 1-[6-methyl-4,5,6,7-tetrahydrothiazolo(5,4-c)pyridin-2-yl] carbonyl-2-carbamoyl-4-(6-chloronaphth-2-ylsulphonyl)piperazine | | Descriptor: | (2R)-4-[(6-CHLORO-2-NAPHTHYL)SULFONYL]-1-[(5-METHYL-4,5,6,7-TETRAHYDRO[1,3]THIAZOLO[5,4-C]PYRIDIN-2-YL)CARBONYL]PIPERAZ INE-2-CARBOXAMIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and conformational analysis of a non-amidine factor Xa inhibitor that incorporates 5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine as S4 binding element
J.Med.Chem., 47, 2004
|
|
2Y6S
 
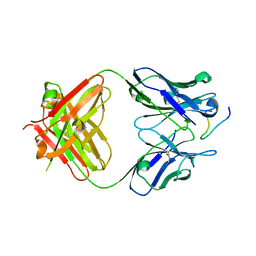 | | Structure of an Ebolavirus-protective antibody in complex with its mucin-domain linear epitope | | Descriptor: | ENVELOPE GLYCOPROTEIN, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Olal, D.O, Kuehne, A, Lee, J.E, Bale, S, Dye, J.M, Saphire, E.O. | | Deposit date: | 2011-01-25 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an Ebola Virus-Protective Antibody in Complex with its Mucin-Domain Linear Epitope.
J.Virol., 86, 2012
|
|
3A5R
 
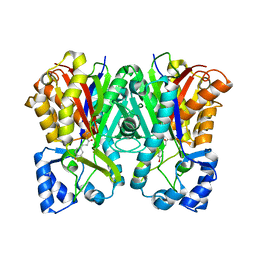 | | Benzalacetone synthase from Rheum palmatum complexed with 4-coumaroyl-primed monoketide intermediate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Benzalacetone synthase | | Authors: | Morita, H, Kato, R, Abe, I, Sugio, S, Kohno, T. | | Deposit date: | 2009-08-10 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structure-based mechanism for benzalacetone synthase from Rheum palmatum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A5Q
 
 | | Benzalacetone synthase from Rheum palmatum | | Descriptor: | Benzalacetone synthase | | Authors: | Morita, H, Kato, R, Abe, I, Sugio, S, Kohno, T. | | Deposit date: | 2009-08-10 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for benzalacetone synthase from Rheum palmatum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A5S
 
 | | Benzalacetone synthase (I207L/L208F) | | Descriptor: | Benzalacetone synthase | | Authors: | Morita, H, Kato, R, Abe, I, Sugio, S, Kohno, T. | | Deposit date: | 2009-08-10 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for benzalacetone synthase from Rheum palmatum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7CBF
 
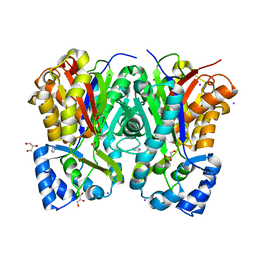 | | Crystal structure of benzophenone synthase from Garcinia mangostana L. pericarps reveals basis for substrate specificity and catalysis | | Descriptor: | 2,4,6-trihydroxybenzophenone synthase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Songsiriritthigul, C, Nualkaew, N, Chen, C.-J. | | Deposit date: | 2020-06-12 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of benzophenone synthase from Garcinia mangostana L. pericarps reveals basis for substrate specificity and catalysis.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1B1Y
 
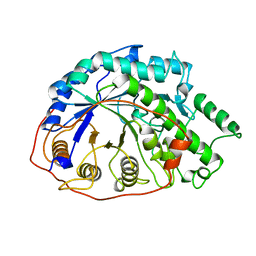 | | SEVENFOLD MUTANT OF BARLEY BETA-AMYLASE | | Descriptor: | PROTEIN (BETA-AMYLASE), alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose | | Authors: | Mikami, B, Yoon, H.J, Yoshigi, N. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the sevenfold mutant of barley beta-amylase with increased thermostability at 2.5 A resolution.
J.Mol.Biol., 285, 1999
|
|
5XXT
 
 | | GDP-microtubule complexed with nucleotide-free KIF5C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | Deposit date: | 2017-07-05 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.35 Å) | | Cite: | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
5XXW
 
 | | GDP-microtubule complexed with KIF5C in ATP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | Deposit date: | 2017-07-05 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
5XXV
 
 | | GDP-microtubule complexed with KIF5C in AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | Deposit date: | 2017-07-05 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (6.46 Å) | | Cite: | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
5XXX
 
 | | GMPCPP-microtubule complexed with nucleotide-free KIF5C | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | Deposit date: | 2017-07-05 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (6.43 Å) | | Cite: | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
