3IJ7
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | 4-O-methyl-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranosyl fluoride, 4-O-methyl-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-2)-5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-03 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3IJ8
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, 5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
1RZ2
 
 | | 1.6A crystal structure of the protein BA4783/Q81L49 (similar to sortase B) from Bacillus anthracis. | | Descriptor: | conserved hypothetical protein BA4783 | | Authors: | Wu, R, Zhang, R, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-23 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site.
Structure, 12, 2004
|
|
3IJ9
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
1SQE
 
 | | 1.5A Crystal Structure Of the protein PG130 from Staphylococcus aureus, Structural genomics | | Descriptor: | hypothetical protein PG130 | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases
J.Biol.Chem., 280, 2005
|
|
1PVM
 
 | | Crystal Structure of a Conserved CBS Domain Protein TA0289 of Unknown Function from Thermoplasma acidophilum | | Descriptor: | MERCURY (II) ION, conserved hypothetical protein Ta0289 | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Xu, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-27 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain
J.Mol.Biol., 375, 2008
|
|
1PC6
 
 | | Structural Genomics, NinB | | Descriptor: | BETA-MERCAPTOETHANOL, Protein ninB | | Authors: | Zhang, R, Beasley, S, Maxwell, K.L, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-15 | | Release date: | 2004-01-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Functional similarities between phage lambda Orf and Escherichia coli RecFOR in initiation of genetic exchange
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6IHB
 
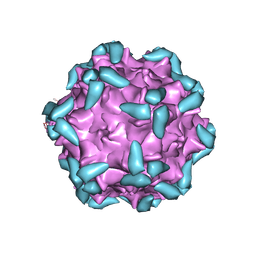 | | Adeno-Associated Virus 2 in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
5Y8E
 
 | |
5Y8F
 
 | |
2R2Q
 
 | | Crystal structure of human Gamma-Aminobutyric Acid Receptor-Associated Protein-like 1 (GABARAP1), Isoform CRA_a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 1, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Paramanathan, R, Davis, T, Mujib, S, Butler-Cole, C, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human Gamma-Aminobutyric Acid Receptor-Associated Protein-like 1 (GABARAP1), Isoform CRA_a.
To be Published
|
|
6IH9
 
 | | Adeno-Associated Virus 2 at 2.8 ang | | Descriptor: | Capsid protein VP1 | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
2QKU
 
 | |
2QKV
 
 | |
1PIN
 
 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Noel, J.P, Ranganathan, R, Hunter, T. | | Deposit date: | 1998-06-21 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
2QKT
 
 | |
6JCS
 
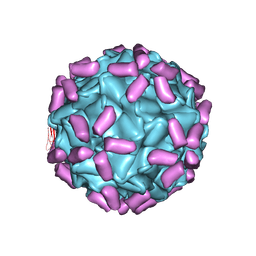 | | AAV5 in complex with AAVR | | Descriptor: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCR
 
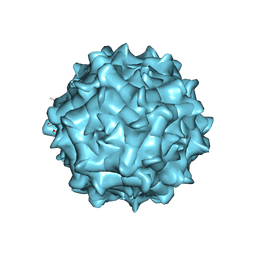 | | AAV1 in neutral condition at 3.07 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCQ
 
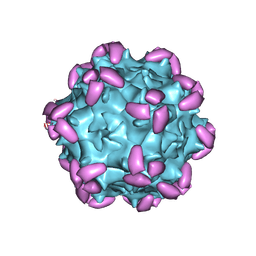 | | AAV1 in complex with AAVR | | Descriptor: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCT
 
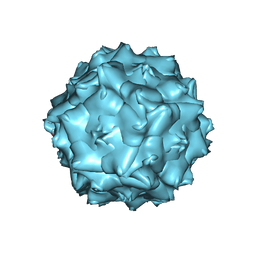 | | AAV5 in neutral condition at 3.18 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-07-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
3T97
 
 | |
6KDU
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6KKV
 
 | |
6KJM
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based identification of first-in-class fragment inhibitors that target the NMN pocket of M. tuberculosis NAD + -dependent DNA ligase A.
J.Struct.Biol., 213, 2021
|
|
6KSD
 
 | |
