6DVD
 
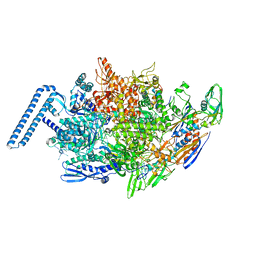 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) with 6 nt spacer and bromine labelled in position "-11 | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*A)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.899 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DV9
 
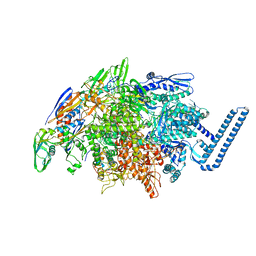 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 4nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVB
 
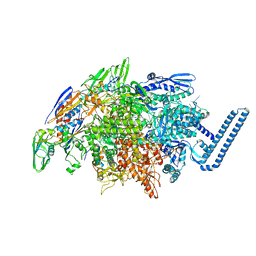 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 5nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVE
 
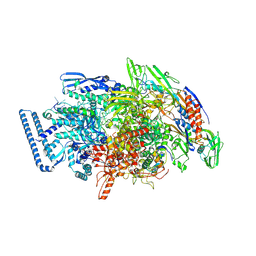 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF selenomethionine-labelled sigma factor L) with 6 nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*GP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVC
 
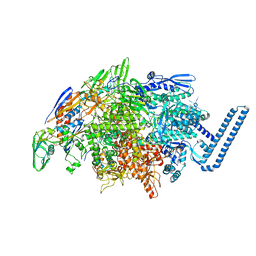 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 6nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
4F20
 
 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Dermokine | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
6IH5
 
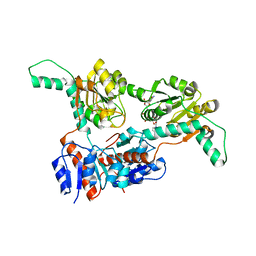 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IQD
 
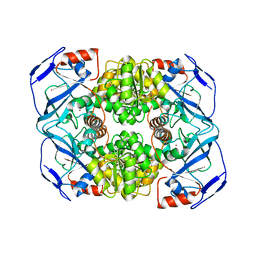 | | Crystal structure of Alcohol dehydrogenase from Geobacillus stearothermophilus | | Descriptor: | Alcohol dehydrogenase, ZINC ION | | Authors: | Xue, S, Feng, Y, Guo, X, Zhao, Z. | | Deposit date: | 2018-11-07 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Characterization of the substrate scope of an alcohol dehydrogenase commonly used as methanol dehydrogenase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6IH8
 
 | |
6IVU
 
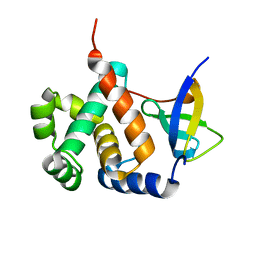 | |
6IVS
 
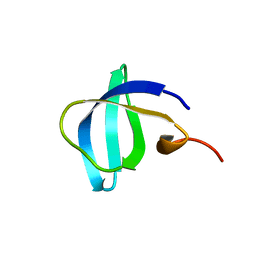 | |
6K4Y
 
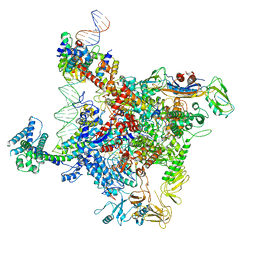 | | CryoEM structure of sigma appropriation complex | | Descriptor: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
6IH6
 
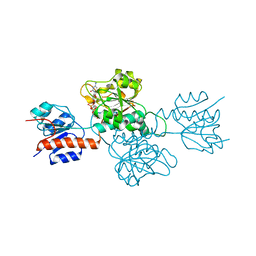 | | Phosphite Dehydrogenase mutant I151R/P176R/M207A from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH3
 
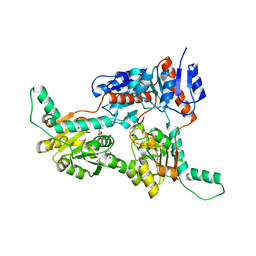 | | Crystal structure of Phosphite Dehydrogenase from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Zhao, Z, Liu, Y. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
 | |
6KJJ
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | 4-(2-acetamidoethylsulfanyl)-4-oxidanylidene-butanoic acid, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJH
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | Putative beta-lactamase, SULFATE ION | | Authors: | Xiao, F, Sheng, D, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJQ
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | (3~{Z},5~{E},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-8,14,16-tris(oxidanyl)-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-2-one, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJR
 
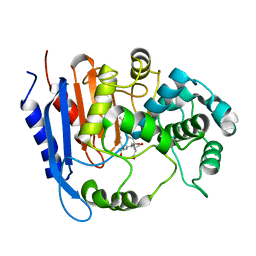 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | 4-[[(3~{E},5~{Z},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-14,16-bis(oxidanyl)-2-oxidanylidene-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-8-yl]oxy]-4-oxidanylidene-butanoic acid, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJP
 
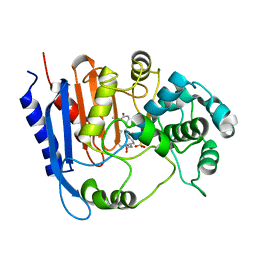 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | (3~{Z},5~{E},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-8,14,16-tris(oxidanyl)-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-2-one, Putative beta-lactamase, SULFATE ION | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJT
 
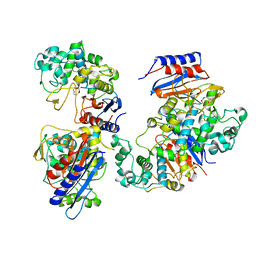 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | Putative beta-lactamase, SUCCINIC ACID | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6IH2
 
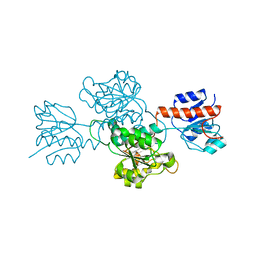 | |
3TN5
 
 | | Crystal structure of GkaP mutant Y99L from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, PHOSPHATE ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
3TN4
 
 | | Crystal structure of GkaP mutant G209D from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
3TN3
 
 | | Crystal structure of GkaP from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
