8PB9
 
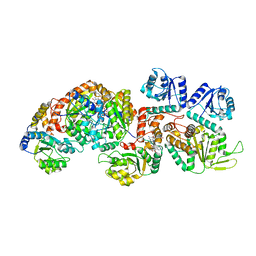 | | Cryo-EM structure of the c-di-GMP-bound FleQ-FleN master regulator complex from Pseudomonas aeruginosa | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Antiactivator FleN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Torres-Sanchez, L.T, Krasteva, P.V. | | Deposit date: | 2023-06-08 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the P. aeruginosa FleQ-FleN master regulators reveal large-scale conformational switching in motility and biofilm control.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8P53
 
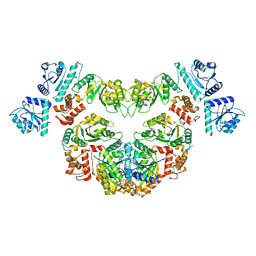 | |
7EJW
 
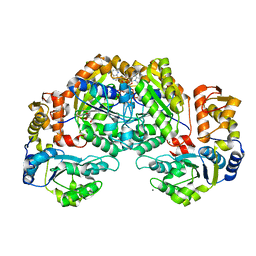 | | Crystal structure of FleN in complex with FleQ AAA+ doamain | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Chanchal, Banerjee, P, Raghav, S, Jain, D. | | Deposit date: | 2021-04-02 | | Release date: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The antiactivator FleN uses an allosteric mechanism to regulate sigma 54 -dependent expression of flagellar genes in Pseudomonas aeruginosa .
Sci Adv, 7, 2021
|
|
6OBY
 
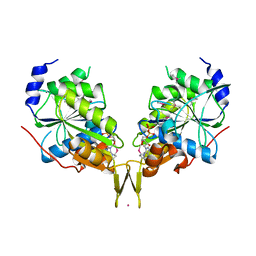 | |
6G2G
 
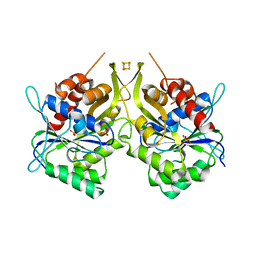 | | Fe-S assembly Cfd1 | | Descriptor: | Cytosolic Fe-S cluster assembly factor CFD1, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2018-03-23 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Function and crystal structure of the dimeric P-loop ATPase CFD1 coordinating an exposed [4Fe-4S] cluster for transfer to apoproteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5J1J
 
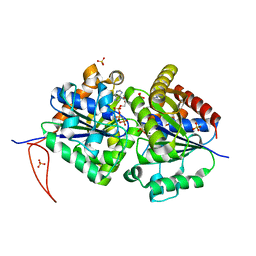 | | Structure of FleN-AMPPNP complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Jain, D, Chanchal, Banerjee, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ATP-Induced Structural Remodeling in the Antiactivator FleN Enables Formation of the Functional Dimeric Form
Structure, 25, 2017
|
|
5JVF
 
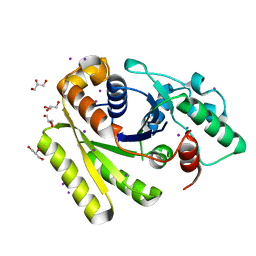 | | Crystal Structure of Apo-FleN | | Descriptor: | GLYCEROL, IODIDE ION, Site-determining protein | | Authors: | Jain, D, Chanchal, Banerjee, P. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | ATP-Induced Structural Remodeling in the Antiactivator FleN Enables Formation of the Functional Dimeric Form
Structure, 25, 2017
|
|
5AUQ
 
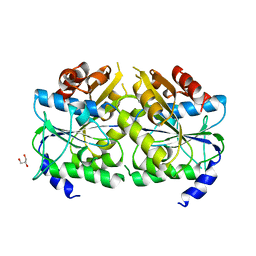 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUP
 
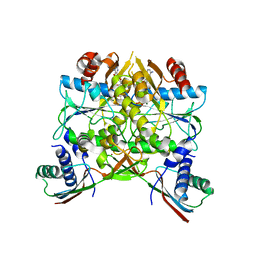 | | Crystal structure of the HypAB complex | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUN
 
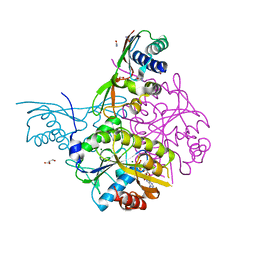 | | Crystal structure of the HypAB-Ni complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ParA/MinD family, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUO
 
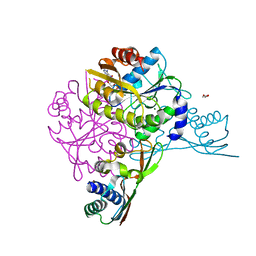 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RZ2
 
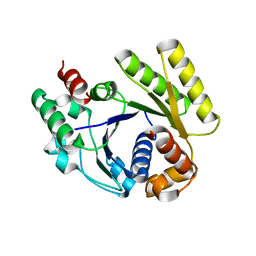 | |
4RZ3
 
 | | Crystal structure of the MinD-like ATPase FlhG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Site-determining protein | | Authors: | Schuhmacher, J.S, Bange, G. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3VX3
 
 | | Crystal structure of [NiFe] hydrogenase maturation protein HypB from Thermococcus kodakarensis KOD1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ... | | Authors: | Sasaki, D, Watanabe, S, Miki, K. | | Deposit date: | 2012-09-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure of a Novel Archaeal HypB for [NiFe] Hydrogenase Maturation
J.Mol.Biol., 425, 2013
|
|
3KB1
 
 | | Crystal Structure of the Nucleotide-binding protein AF_226 in complex with ADP from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR157 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleotide-binding protein, ZINC ION | | Authors: | Forouhar, F, Lew, S, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR157
To be Published
|
|
2PH1
 
 | | Crystal structure of nucleotide-binding protein AF2382 from Archaeoglobus fulgidus, Northeast Structural Genomics Target GR165 | | Descriptor: | Nucleotide-binding protein, ZINC ION | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-04-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of nucleotide-binding protein AF2382 from Archaeoglobus fulgidus.
To be Published
|
|
