7PU7
 
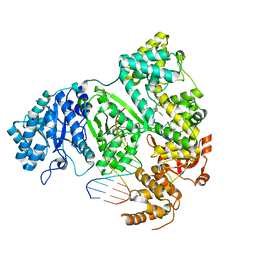 | | DNA polymerase from M. tuberculosis | | Descriptor: | DNA polymerase III subunit alpha, Template, ZINC ION, ... | | Authors: | Borsellini, A, Lamers, M.H. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-23 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | DNA-Dependent Binding of Nargenicin to DnaE1 Inhibits Replication in Mycobacterium tuberculosis.
Acs Infect Dis., 8, 2022
|
|
5LEW
 
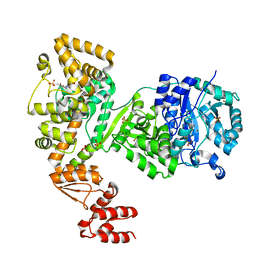 | | DNA polymerase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA polymerase III subunit alpha, SULFATE ION, ... | | Authors: | Banos-Mateos, S, Lang, U.F, Maslen, S.L, Skehel, J.M, Lamers, M.H. | | Deposit date: | 2016-06-30 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-fidelity DNA replication in Mycobacterium tuberculosis relies on a trinuclear zinc center.
Nat Commun, 8, 2017
|
|
5M1S
 
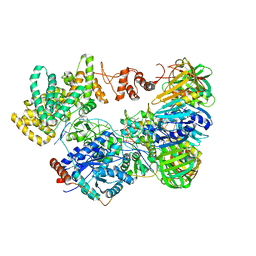 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | Descriptor: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-18 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4IQJ
 
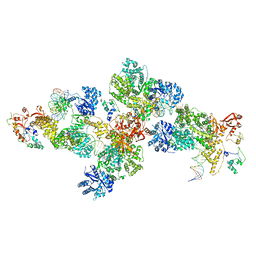 | | Structure of PolIIIalpha-Tauc-DNA complex suggests an atomic model of the replisome | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*GP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*A)-3'), DNA (5'-D(P*CP*GP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(DOC))-3'), ... | | Authors: | Liu, B, Lin, J, Steitz, T. | | Deposit date: | 2013-01-11 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of PolIIIalpha-Tauc-DNA complex suggests an atomic model of the replisome
Structure, 21, 2013
|
|
4JOM
 
 | | Structure of E. coli Pol III 3mPHP mutant | | Descriptor: | DNA polymerase III subunit alpha, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Barros, T, Guenther, J, Kelch, B, Anaya, J, Prabhakar, A, O'Donnell, M, Kuriyan, J, Lamers, M.H. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structural role for the PHP domain in E. coli DNA polymerase III.
Bmc Struct.Biol., 13, 2013
|
|
5FKU
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2019-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKW
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2019-02-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKV
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2019-02-27 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
2HNH
 
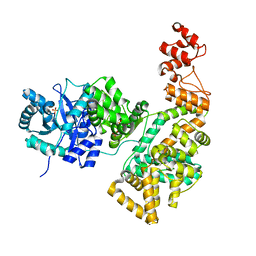 | | Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III | | Descriptor: | DNA polymerase III alpha subunit, PHOSPHATE ION | | Authors: | Meindert, M.H, Georgescu, R.E, Lee, S, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HPI
 
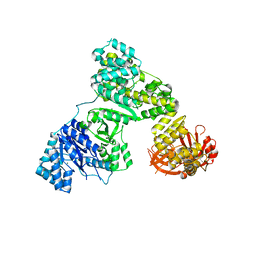 | | Eubacterial and Eukaryotic Replicative DNA Polymerases are not Homologous: X-ray Structure of DNA Polymerase III | | Descriptor: | CHLORIDE ION, DNA polymerase III alpha subunit, MAGNESIUM ION, ... | | Authors: | Bailey, S, Wing, R.A, Steitz, T.A. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of T. aquaticus DNA Polymerase III Is Distinct from Eukaryotic Replicative DNA Polymerases.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HPM
 
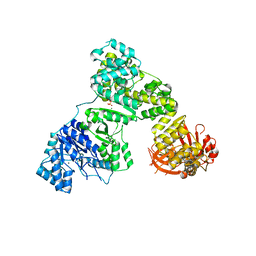 | | Eubacterial and Eukaryotic Replicative DNA Polymerases are not Homologous: X-ray Structure of DNA Polymerase III | | Descriptor: | CHLORIDE ION, DNA Polymerase III alpha subunit, MAGNESIUM ION, ... | | Authors: | Bailey, S, Wing, R.A, Steitz, T.A. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The Structure of T. aquaticus DNA Polymerase III Is Distinct from Eukaryotic Replicative DNA Polymerases.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HQA
 
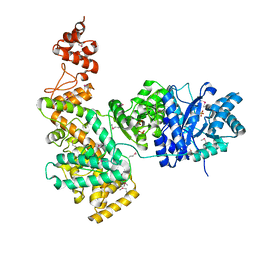 | | Crystal structure of the catalytic alpha subunit of E. Coli replicative DNA polymerase III | | Descriptor: | DNA polymerase III alpha subunit, PHOSPHATE ION | | Authors: | Lamers, M.H, Georgescu, R.E, Lee, S.G, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
3E0D
 
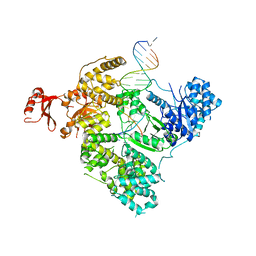 | | Insights into the Replisome from the Crystral Structure of the Ternary Complex of the Eubacterial DNA Polymerase III alpha-subunit | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase III subunit alpha, ... | | Authors: | Wing, R.A, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Insights into the Replisome from the Structure of a Ternary Complex of the DNA Polymerase III alpha-Subunit.
J.Mol.Biol., 382, 2008
|
|
3F2C
 
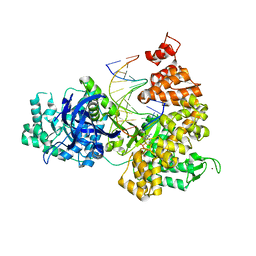 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP and Mn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3F2D
 
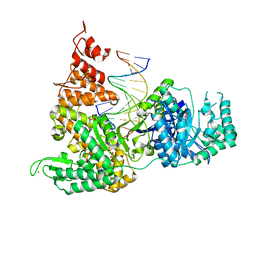 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP, Mn and Zn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3F2B
 
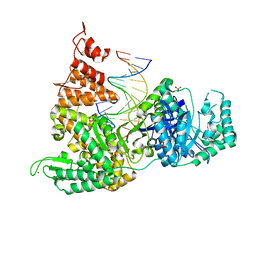 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP, Mg and Zn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
