7AGL
 
 | |
3CZX
 
 | |
1JWQ
 
 | | Structure of the catalytic domain of CwlV, N-acetylmuramoyl-L-alanine amidase from Bacillus(Paenibacillus) polymyxa var.colistinus | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE CwlV, ZINC ION | | Authors: | Yamane, T, Koyama, Y, Nojiri, Y, Hikage, T, Akita, M, Suzuki, A, Shirai, T, Ise, F, Shida, T, Sekiguchi, J. | | Deposit date: | 2001-09-05 | | Release date: | 2003-11-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the catalytic domain of N-acetylmuramoyl-L-alanine amidase, a cell wall hydrolase from Bacillus polymyxa var.colistinus and its resemblance to the structure of carboxypeptidases
To be Published
|
|
7RAG
 
 | |
5EMI
 
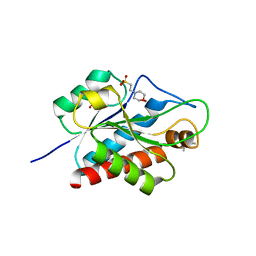 | | N-acetylmuramoyl-L-alanine amidase AmiC2 of Nostoc punctiforme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cell wall hydrolase/autolysin, ... | | Authors: | Buettner, F.M, Stehle, T. | | Deposit date: | 2015-11-06 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Enabling cell-cell communication via nanopore formation: structure, function and localization of the unique cell wall amidase AmiC2 of Nostoc punctiforme.
Febs J., 283, 2016
|
|
4RN7
 
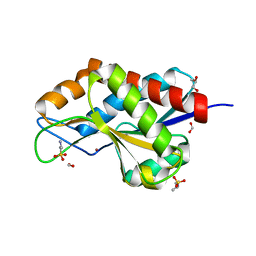 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
7TJ4
 
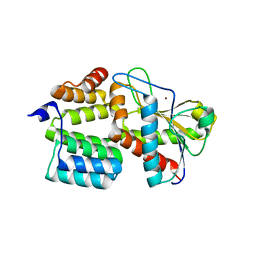 | | Structure of the S. aureus amidase LytH and activator ActH extracellular domains | | Descriptor: | ActH, LytH, ZINC ION | | Authors: | Page, J.E, Skiba, M.A, Kruse, A.C, Walker, S. | | Deposit date: | 2022-01-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metal cofactor stabilization by a partner protein is a widespread strategy employed for amidase activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7B3N
 
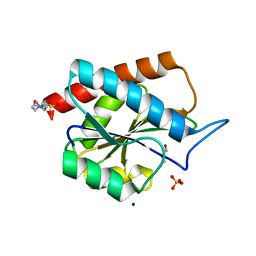 | | AmiP amidase-3 from Thermus parvatiensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cell wall hydrolase, ... | | Authors: | Freitag-Pohl, S, Pohl, E. | | Deposit date: | 2020-12-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | AmiP from hyperthermophilic Thermus parvatiensis prophage is a thermoactive and ultrathermostable peptidoglycan lytic amidase.
Protein Sci., 32, 2023
|
|
3NE8
 
 | | The crystal structure of a domain from N-acetylmuramoyl-l-alanine amidase of Bartonella henselae str. Houston-1 | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-14 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division.
Mol.Microbiol., 85, 2012
|
|
8C2O
 
 | | Structure of E. coli AmiA | | Descriptor: | N-acetylmuramoyl-L-alanine amidase AmiA, ZINC ION | | Authors: | Baverstock, T.C, Crow, A. | | Deposit date: | 2022-12-22 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Activator-induced conformational changes regulate division-associated peptidoglycan amidases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7AGO
 
 | | crystal structure of the N-acetylmuramyl-L-alanine amidase, Ami1, from Mycobacterium abscessus bound to L-Alanine-D-isoglutamine | | Descriptor: | ALANINE, D-alpha-glutamine, N-acetylmuramoyl-L-alanine amidase, ... | | Authors: | Blaise, M. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Characterization of the N -Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus .
Cells, 9, 2020
|
|
7AGM
 
 | | Crystal structure of the N-acetylmuramyl-L-alanine amidase, Ami1, from Mycobacterium smegmatis | | Descriptor: | N-acetylmuramoyl-L-alanine amidase, ZINC ION | | Authors: | Blaise, M, Alsarraf, M.A.B. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Functional Characterization of the N -Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus .
Cells, 9, 2020
|
|
3QAY
 
 | | Catalytic domain of CD27L endolysin targeting Clostridia Difficile | | Descriptor: | Endolysin, PHOSPHATE ION, ZINC ION | | Authors: | Mayer, M.J, Garefaliki, V, Spoerl, R, Narbad, A, Meijers, R. | | Deposit date: | 2011-01-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range.
J.Bacteriol., 193, 2011
|
|
4LQ6
 
 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4M6H
 
 | | Structure of the reduced, metal-free form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717 | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4M6G
 
 | | Structure of the Mycobacterium tuberculosis peptidoglycan amidase Rv3717 in complex with L-Alanine-iso-D-Glutamine reaction product | | Descriptor: | ALANINE, D-alpha-glutamine, Peptidoglycan Amidase Rv3717, ... | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4M6I
 
 | | Structure of the reduced, Zn-bound form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717, ZINC ION | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
5J72
 
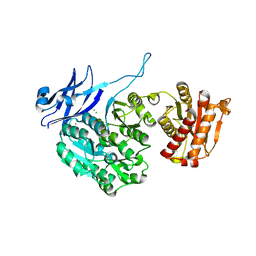 | | Cwp6 from Clostridium difficile | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
1XOV
 
 | |
4BIN
 
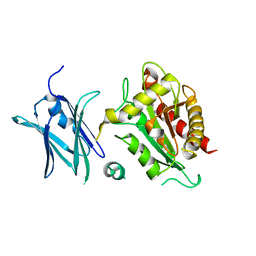 | | Crystal structure of the E. coli N-acetylmuramoyl-L-alanine amidase AmiC | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE AMIC, SODIUM ION, ZINC ION | | Authors: | Kerff, F, Rocaboy, M, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2013-04-12 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Crystal Structure of the Cell Division Amidase Amic Reveals the Fold of the Amin Domain, a New Peptidoglycan Binding Domain.
Mol.Microbiol., 90, 2013
|
|
8C0J
 
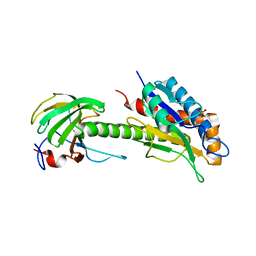 | | Structure of AmiB enzymatic domain bound to the EnvC LytM domain | | Descriptor: | Murein hydrolase activator EnvC, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Crow, A. | | Deposit date: | 2022-12-17 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.381 Å) | | Cite: | Activator-induced conformational changes regulate division-associated peptidoglycan amidases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
