8ZAN
 
 | | ExoChR2 channelrhodopsin | | Descriptor: | Archaeal-type opsin 2, RETINAL | | Authors: | Zhang, M.F. | | Deposit date: | 2024-04-25 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Channelrhodopsins with distinct chromophores and binding patterns.
Nat Commun, 15, 2024
|
|
8YEK
 
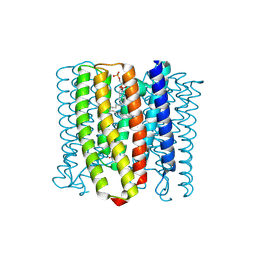 | | Cryo-EM structure of the channelrhodopsin GtCCR2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 2024
|
|
8YEJ
 
 | | Cryo-EM structure of the channelrhodopsin GtCCR2 focused on the monomer | | Descriptor: | GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 2024
|
|
8YEL
 
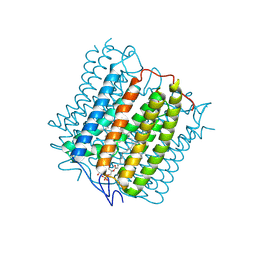 | | Cryo-EM structure of the channelrhodopsin GtCCR4 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cation channel rhodopsin 4, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 2024
|
|
8QQZ
 
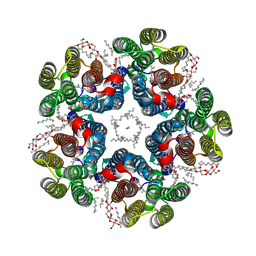 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QR0
 
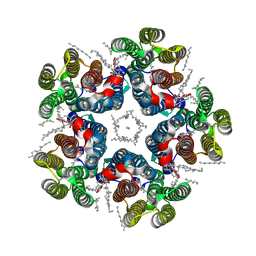 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QQO
 
 | |
8QLE
 
 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 4.6 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QLF
 
 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 8.8 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8JH0
 
 | | Crystal structure of the light-driven sodium pump IaNaR | | Descriptor: | RETINAL, Xanthorhodopsin | | Authors: | Hashimoto, T, Kato, K, Tanaka, Y, Yao, M, Kikukawa, T. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Multistep conformational changes leading to the gate opening of light-driven sodium pump rhodopsin.
J.Biol.Chem., 299, 2023
|
|
8CQC
 
 | |
8CQD
 
 | |
8CNK
 
 | |
8CL7
 
 | | Krokinobacter eikastus rhodopsin 2 (KR2) in dark state | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Bertrand, Q, Wranik, M, Kepa, M.W, Weinert, T, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
8CL8
 
 | | Krokinobacter eikastus rhodopsin 2 (KR2) extrapolated map 1us after light activation | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Bertrand, Q, Kepa, M.W, Weinert, T, Wranik, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
8I2Z
 
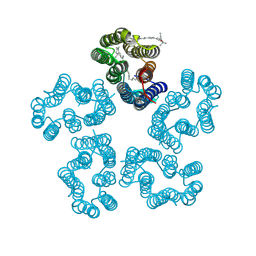 | | Cryo-EM structure of the zeaxanthin-bound kin4B8 | | Descriptor: | RETINAL, Xanthorhodopsin, Zeaxanthin | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2023-01-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
7YTB
 
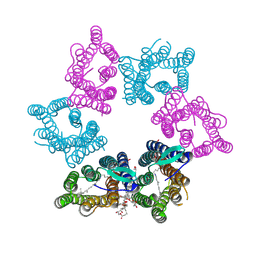 | | Crystal structure of Kin4B8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kin4B8, RETINAL | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
8ANQ
 
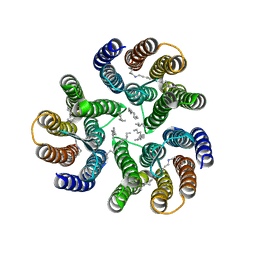 | | Crystal structure of the microbial rhodopsin from Sphingomonas paucimobilis (SpaR) | | Descriptor: | Bacteriorhodopsin, EICOSANE | | Authors: | Kovalev, K, Okhrimenko, I, Marin, E, Gordeliy, V. | | Deposit date: | 2022-08-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mirror proteorhodopsins.
Commun Chem, 6, 2023
|
|
7ZY3
 
 | | Room temperature structure of Archaerhodopsin-3 obtained 110 ns after photoexcitation | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kwan, T.O.C, Judge, P.J, Moraes, I, Watts, A, Axford, D, Bada Juarez, J.F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
7XJC
 
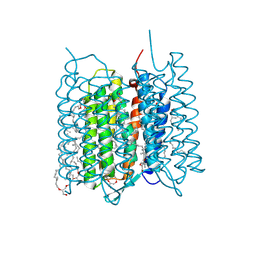 | | Crystal structure of bacteriorhodopsin in the ground and K states after green laser irradiation | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Commun Biol, 6, 2023
|
|
7XJE
 
 | | Crystal structure of bacteriorhodopsin in the K state refined against the extrapolated dataset | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Commun Biol, 6, 2023
|
|
7XJD
 
 | | Crystal structure of bacteriorhodopsin in the ground state by red laser irradiation | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin.
Commun Biol, 6, 2023
|
|
7ZCM
 
 | |
7Z09
 
 | | Crystal structure of the ground state of bacteriorhodopsin at 1.05 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0A
 
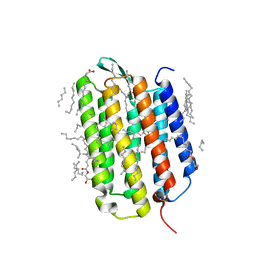 | | Crystal structure of the ground state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
