2XRY
 
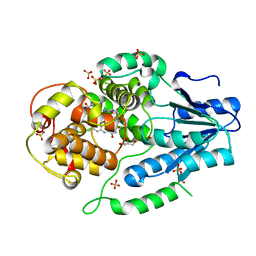 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
7F8T
 
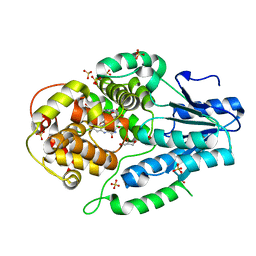 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
3UMV
 
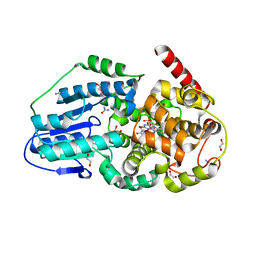 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2011-11-14 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
2XRZ
 
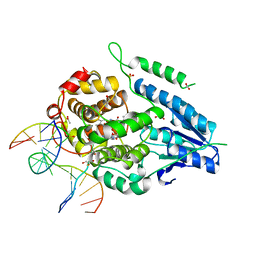 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei in complex with intact CPD-lesion | | Descriptor: | ACETATE ION, COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
5ZCW
 
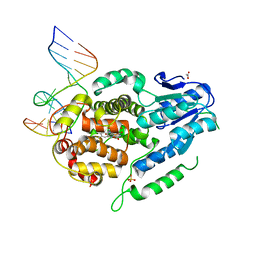 | | Structure of the Methanosarcina mazei class II CPD-photolyase in complex with intact, phosphodiester linked, CPD-lesion | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*(TTD)P*CP*GP*CP*GP*CP*AP*A)-3', 5'-D(*TP*GP*CP*GP*CP*GP*AP*AP*GP*CP*CP*GP*AP*T)-3', ACETATE ION, ... | | Authors: | Maestre-Reyna, M, Bessho, Y. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA.
IUCrJ, 5, 2018
|
|
4CDM
 
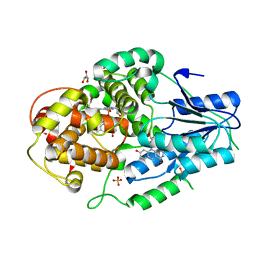 | | Crystal structure of M. mazei photolyase soaked with synthetic 8-HDF | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
4CDN
 
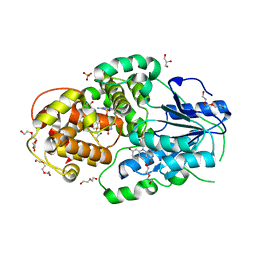 | | Crystal structure of M. mazei photolyase with its in vivo reconstituted 8-HDF antenna chromophore | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
5O86
 
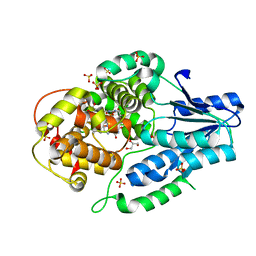 | |
5O8E
 
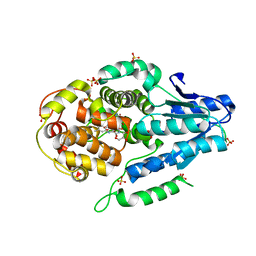 | | Mutant of class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ignatz, E, Geisselbrecht, Y, Essen, L.-O. | | Deposit date: | 2017-06-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nicotinamide Adenine Dinucleotides Arrest Photoreduction of Class II DNA Photolyases in FADH ̇ State.
Photochem. Photobiol., 94, 2018
|
|
5O8D
 
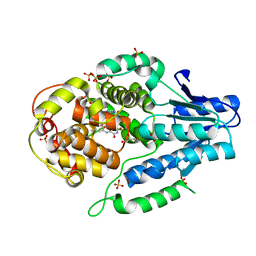 | |
8OET
 
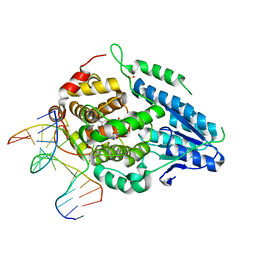 | | SFX structure of the class II photolyase complexed with a thymine dimer | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, DNA (14-mer), Deoxyribodipyrimidine photo-lyase, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-03-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
8OYC
 
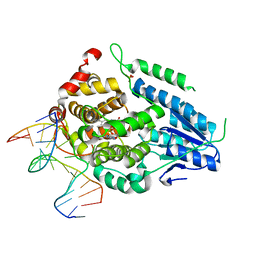 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (100 microsecond timpeoint) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY5
 
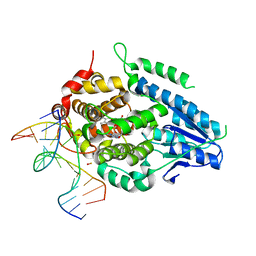 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (1 nanosecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY8
 
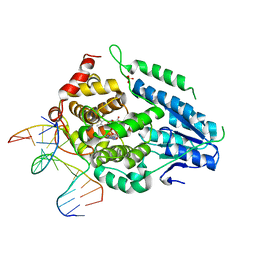 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (30 nanosecond timepoint) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY9
 
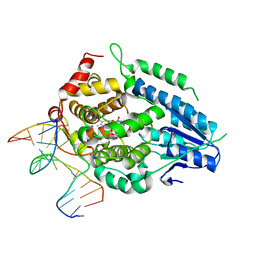 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (1 microsecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY6
 
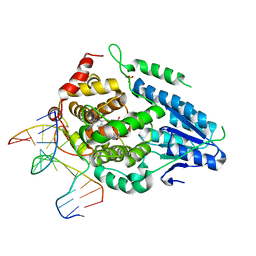 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (3 nanosecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OYA
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (10 microsecond pump probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY3
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (3 picosecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY7
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (10 nanosecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OYB
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (30 microsecond pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8OY4
 
 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (300 ps pump-probe delay) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
7VJ6
 
 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the semiquinone state | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJI
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (10 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ4
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (1 ms time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
