9UUU
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae reduced by NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-05-08 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD9
 
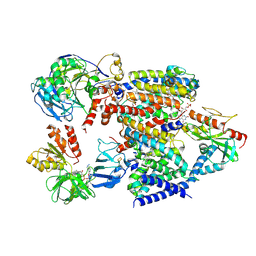 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae reduced by NADH, in the absence of Na+, down state | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-07 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD2
 
 | |
9UD3
 
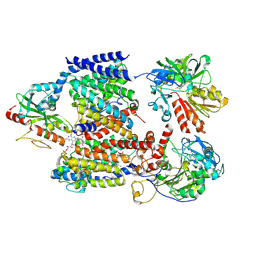 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-T236Y mutant from Vibrio cholerae | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD4
 
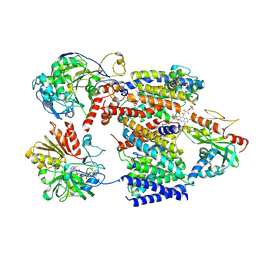 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-T236Y mutant from Vibrio cholerae reduced by NADH | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD6
 
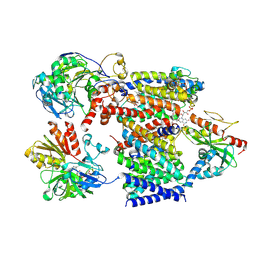 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae reduced by NADH, in the absence of Na+, upper state | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UDA
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae reduced by NADH, with bound korormicin A, stable state | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UD5
 
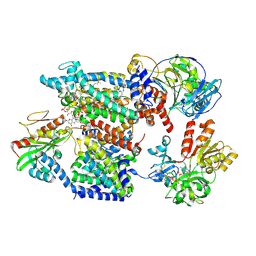 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae reduced by NADH, with bound korormicin A | | Descriptor: | CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UDF
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae reduced by NADH, with bound korormicin A, shifted state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9UDG
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase from Vibrio cholerae reduced by NADH, with bound aurachin D-42 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aurachin D, CALCIUM ION, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9U5G
 
 | |
9LRR
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae with bound korormicin A | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-02-01 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural Elucidation of the Mechanism for Inhibitor Resistance in the Na + -Translocating NADH-Ubiquinone Oxidoreductase from Vibrio cholerae.
Biochemistry, 64, 2025
|
|
9I2A
 
 | |
9HI3
 
 | |
9GYU
 
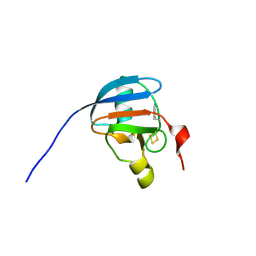 | | Ferredoxin CNF-labelled reduced state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-03 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 2025
|
|
9GYL
 
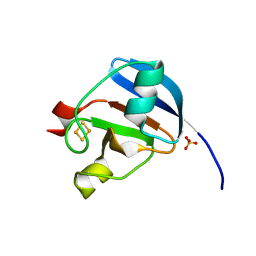 | | Ferredoxin Wild-type -Oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic, ... | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 2025
|
|
9GYR
 
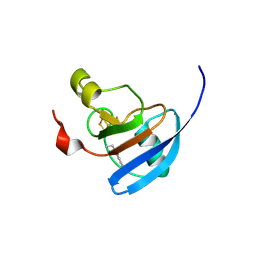 | | Ferredoxin CNF labelled, oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 2025
|
|
9GYN
 
 | | Ferredoxin Wild-type - Reduced state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 2025
|
|
9GYD
 
 | | Ferredoxin Wild-type -As-isolated state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-01 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 2025
|
|
9GRX
 
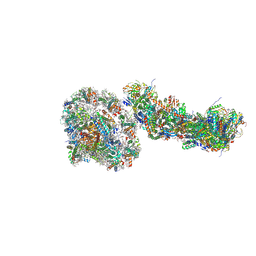 | | NDH-PSI-LHCI supercomplex from S. oleracea | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Introini, B, Hahn, A, Kuehlbrandt, W. | | Deposit date: | 2024-09-13 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure of the NDH-PSI-LHCI supercomplex from Spinacia oleracea.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9CU0
 
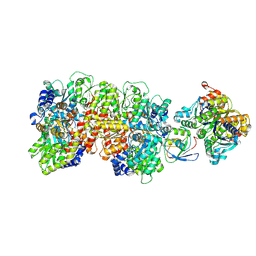 | | Azotobacter vinelandii 1:1:1 MoFeP:FeP:FeSII-Complex (C1 symmetry) | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Narehood, S.M, Cook, B.D, Srisantitham, S, Eng, V.H, Shiau, A, Britt, R.D, Herzik, M.A, Tezcan, F.A. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis for the conformational protection of nitrogenase from O 2 .
Nature, 637, 2025
|
|
9CU2
 
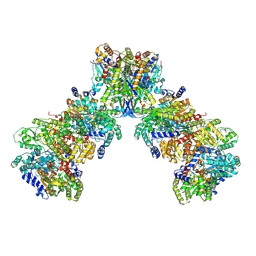 | | Azotobacter vinelandii filamentous 2:2:1 MoFeP:FeP:FeSII-Complex (C2 symmetry) | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Narehood, S.M, Cook, B.D, Srisantitham, S, Eng, V.H, Shiau, A, Britt, R.D, Herzik, M.A, Tezcan, F.A. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Structural basis for the conformational protection of nitrogenase from O 2 .
Nature, 637, 2025
|
|
9CU1
 
 | | Azotobacter vinelandii filamentous 2:2:1 MoFeP:FeP:FeSII-Complex (termini; C1 symmetry) | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Narehood, S.M, Cook, B.D, Srisantitham, S, Eng, V.H, Shiau, A, Britt, R.D, Herzik, M.A, Tezcan, F.A. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for the conformational protection of nitrogenase from O 2 .
Nature, 637, 2025
|
|
8RMG
 
 | |
8RMF
 
 | |
