3GZC
 
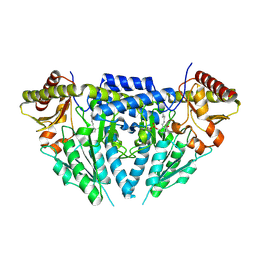 | | Structure of human selenocysteine lyase | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Selenocysteine lyase | | Authors: | Collins, R, Hogbom, M, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
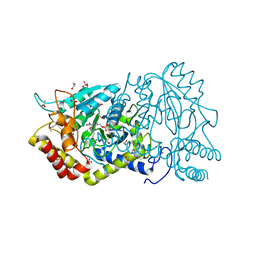
3KE3
 
 | |
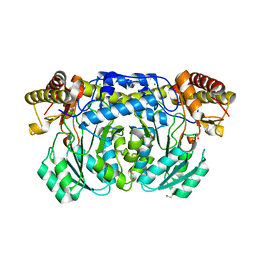
3KGW
 
 | |
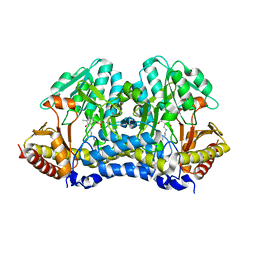
4Q76
 
 | |
4Q75
 
 | |
3GZD
 
 | | Human selenocysteine lyase, P1 crystal form | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, NITRATE ION, Selenocysteine lyase | | Authors: | Karlberg, T, Hogbom, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
4R5F
 
 | | X-ray structure of the D199K mutant of the cysteine desulfurase IscS from A. fulgidus | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETATE ION, CALCIUM ION, ... | | Authors: | Pagnier, A, Nicolet, Y, Fontecilla-Camps, J.C. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IscS from Archaeoglobus fulgidus has no desulfurase activity but may provide a cysteine ligand for [Fe2S2] cluster assembly.
Biochim.Biophys.Acta, 1853, 2015
|
|
5HHY
 
 | | Structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) showing X-Ray induced reduction of PLP internal aldimine to 4'-deoxy-piridoxine-phosphate (PLR) | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
1VJO
 
 | |
3ISL
 
 | | Crystal structure of ureidoglycine-glyoxylate aminotransferase (pucG) from Bacillus subtilis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Purine catabolism protein pucG | | Authors: | Costa, R, Cendron, L, Ramazzina, I, Berni, R, Peracchi, A, Percudani, R, Zanotti, G. | | Deposit date: | 2009-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Amino acids from purines in GUT bacteria
To be Published
|
|
1W23
 
 | | Crystal structure of phosphoserine aminotransferase from Bacillus alcalophilus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dubnovitsky, A, Kapetaniou, E.G, Papageorgiou, A.C. | | Deposit date: | 2004-06-25 | | Release date: | 2004-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Enzyme Adaptation to Alkaline Ph: Atomic Resolution (1.08 A) Structure of Phosphoserine Aminotransferase from Bacillus Alcalophilus
Protein Sci., 14, 2005
|
|
1BT4
 
 | |
1W3U
 
 | |
1BJO
 
 | |
5FT5
 
 | | Crystal structure of the cysteine desulfurase CsdA (persulfurated) from Escherichia coli at 2.384 Angstroem resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
5J8Q
 
 | |
1C0N
 
 | | CSDB PROTEIN, NIFS HOMOLOGUE | | Descriptor: | ACETIC ACID, PROTEIN (CSDB PROTEIN), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Fujii, T, Maeda, M, Mihara, H, Kurihara, T, Esaki, N, Hata, Y. | | Deposit date: | 1999-07-17 | | Release date: | 2000-07-17 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase
Biochemistry, 39, 2000
|
|
5LUC
 
 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.8 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Borri Voltattorni, C, Montioli, R. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
1N31
 
 | | Structure of A Catalytically Inactive Mutant (K223A) of C-DES with a Substrate (Cystine) Linked to the Co-Factor | | Descriptor: | CYSTEINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
6PK3
 
 | |
6CZY
 
 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with Pyridoxamine-5'-phosphate (PMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
6PK1
 
 | |
6CZX
 
 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP internal aldimine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
6CZZ
 
 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP-phosphoserine geminal diamine intermediate | | Descriptor: | PHOSPHOSERINE, PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
1N2T
 
 | | C-DES Mutant K223A with GLY Covalenty Linked to the PLP-cofactor | | Descriptor: | GLYCINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-24 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
