9GZR
 
 | |
9H6U
 
 | | SARS-CoV-2 S protein in complex with pT1679 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, pT1679 Fab heavy chain, ... | | Authors: | Hansen, G, Benecke, T, Vollmer, B, Gruenewald, K, Krey, T. | | Deposit date: | 2024-10-25 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | A critical residue in a conserved RBD epitope determines neutralization breadth of pan-sarbecovirus antibodies with recurring YYDRxxG motifs.
Mbio, 2025
|
|
9H7X
 
 | | SmsC2 dimer from M. jannaschii | | Descriptor: | AMP PHOSPHORAMIDATE, Uncharacterized ABC transporter ATP-binding protein MJ0035, ZINC ION | | Authors: | Mechaly, A.E, Dussouchaud, M, Haouz, A, Betton, J.M, Barras, F. | | Deposit date: | 2024-10-28 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Ancestral [Fe-S] biogenesis system SMS has a unique mechanism of cluster assembly and sulfur utilization.
Plos Biol., 23, 2025
|
|
9H7Y
 
 | | SmsC2B2 complex from M. jannaschii (orthorhombic form) | | Descriptor: | Iron-sulfur cluster assembly SufBD family protein MJ0034, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Mechaly, A.E, Dussouchaud, M, Haouz, A, Betton, J.M, Barras, F. | | Deposit date: | 2024-10-28 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Ancestral [Fe-S] biogenesis system SMS has a unique mechanism of cluster assembly and sulfur utilization.
Plos Biol., 23, 2025
|
|
9HBL
 
 | | SmsC2B2 complex from M. jannaschii (monoclinic form) | | Descriptor: | 1,2-ETHANEDIOL, Iron-sulfur cluster assembly SufBD family protein MJ0034, SULFATE ION, ... | | Authors: | Mechaly, A.E, Dussouchaud, M, Haouz, A, Betton, J.M, Barras, F. | | Deposit date: | 2024-11-07 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Ancestral [Fe-S] biogenesis system SMS has a unique mechanism of cluster assembly and sulfur utilization.
Plos Biol., 23, 2025
|
|
9HEH
 
 | | Structure of the ligand binding domain of the chemoreceptor MkcA (DSOMK10_RS0100305) of Dickeya solani MK10 in complex with choline | | Descriptor: | 1,2-ETHANEDIOL, CHOLINE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Matilla, M.A. | | Deposit date: | 2024-11-14 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Acetylcholine chemotaxis in global bacterial plant pathogens.
Microbiol Res, 300, 2025
|
|
9HML
 
 | | KIR2DL1 bound to RIFIN PfKE01_040007400 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, KIR2DL protein, RIFIN PfKE01_040007400, ... | | Authors: | Chamberlain, S.G, Higgins, M.K. | | Deposit date: | 2024-12-09 | | Release date: | 2025-04-09 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | RIFINs displayed on malaria-infected erythrocytes bind KIR2DL1 and KIR2DS1.
Nature, 643, 2025
|
|
9HVB
 
 | | High-efficiency Kemp eliminases by complete computational design | | Descriptor: | Kemp eliminase, SULFATE ION | | Authors: | Listov, D, Dym, O, Vos, E, Hoffka, G, Berg, A, Rogotner, S, Caroline, s, Kamerlin, L, Fleishman, S.J. | | Deposit date: | 2024-12-26 | | Release date: | 2025-06-11 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complete computational design of high-efficiency Kemp elimination enzymes.
Nature, 643, 2025
|
|
9HVG
 
 | | High-efficiency Kemp eliminases by complete computational design | | Descriptor: | Kemp Elimination | | Authors: | Listov, D, Dym, O, Vos, E, Hoffka, G, Berg, A, Rogotner, S, Caroline, s, Kamerlin, L, Fleishman, S.J. | | Deposit date: | 2024-12-29 | | Release date: | 2025-06-11 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Complete computational design of high-efficiency Kemp elimination enzymes.
Nature, 643, 2025
|
|
9HVH
 
 | | High-efficiency Kemp eliminases by complete computational design | | Descriptor: | KEMP Eliminase | | Authors: | Listov, D, Dym, O, Vos, E, Hoffka, G, Berg, A, Rogotner, S, Caroline, s, Kamerlin, L, Fleishman, S.J. | | Deposit date: | 2024-12-29 | | Release date: | 2025-06-11 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complete computational design of high-efficiency Kemp elimination enzymes.
Nature, 643, 2025
|
|
9I51
 
 | | Crystal structure of the SARS-CoV-2 helicase NSP13 in complex with ADP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Kloskowski, P, Neumann, P, Ficner, R. | | Deposit date: | 2025-01-27 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Nucleotide-bound crystal structures of the SARS-CoV-2 helicase NSP13.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9I53
 
 | | Crystal structure of the SARS-CoV-2 helicase NSP13 in complex with ATP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Kloskowski, P, Neumann, P, Ficner, R. | | Deposit date: | 2025-01-27 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Nucleotide-bound crystal structures of the SARS-CoV-2 helicase NSP13.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9IGQ
 
 | | Crystal structure of PPK2 class III from Erysipelotrichaceae bacterium in complex with AppCH2p and polyphosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, GLYCEROL, ... | | Authors: | Rasche, R, Lawrence-Doerner, A.-M, Cornelissen, N.V, Kuemmel, D. | | Deposit date: | 2025-02-20 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided engineering of a polyphosphate kinase 2 class III from an Erysipelotrichaceae bacterium to produce base-modified purine nucleotides.
Rsc Chem Biol, 6, 2025
|
|
9IGR
 
 | | Crystal structure of PPK2 class III from Erysipelotrichaceae bacterium in complex with polyphosphate | | Descriptor: | BENZOIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rasche, R, Lawrence-Doerner, A.-M, Cornelissen, N.V, Kuemmel, D. | | Deposit date: | 2025-02-20 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Structure-guided engineering of a polyphosphate kinase 2 class III from an Erysipelotrichaceae bacterium to produce base-modified purine nucleotides.
Rsc Chem Biol, 6, 2025
|
|
9IH6
 
 | | A-SPECTRIN SH3 DOMAIN V9Y, A11V, V23L, M25V, V53L, V58L MUTANT | | Descriptor: | GLYCEROL, Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Vizarraga, D, Fita, I, Cianferoni, D, Delgado, J, Serrano, L. | | Deposit date: | 2025-02-20 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Artificial intelligence and first-principle methods in protein redesign: A marriage of convenience?
Protein Sci., 34, 2025
|
|
9IH7
 
 | | A-SPECTRIN SH3 DOMAIN V9L, A11V, V23I, M25V, V46F, V53L, V58L MUTANT | | Descriptor: | SULFATE ION, Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Vizarraga, D, Fita, I, Cianferoni, D, Delgado, J, Serrano, L. | | Deposit date: | 2025-02-20 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Artificial intelligence and first-principle methods in protein redesign: A marriage of convenience?
Protein Sci., 34, 2025
|
|
9IHA
 
 | | A-SPECTRIN SH3 DOMAIN V9L, A11V, M25V, V46F, V58L MUTANT | | Descriptor: | SULFATE ION, Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Vizarraga, D, Fita, I, Cianferoni, D, Delgado, J, Serrano, L. | | Deposit date: | 2025-02-20 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.013 Å) | | Cite: | Artificial intelligence and first-principle methods in protein redesign: A marriage of convenience?
Protein Sci., 34, 2025
|
|
9II2
 
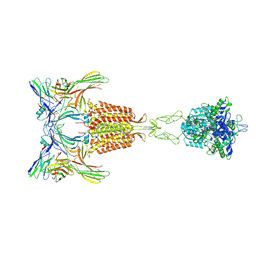 | | Cryo-EM Structure of the 2:2 Complex of mGlu3 and beta-arrestin1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-arrestin-1, CHOLESTEROL, ... | | Authors: | Wen, T.L, Du, M, Yang, X, Shen, Y.Q. | | Deposit date: | 2024-06-18 | | Release date: | 2025-03-05 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to the metabotropic glutamate receptor mGlu3.
Nat.Chem.Biol., 21, 2025
|
|
9II3
 
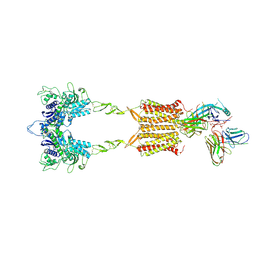 | | Cryo-EM Structure of the 2:1 Complex of mGlu3 and beta-arrestin1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-arrestin-1, CHOLESTEROL, ... | | Authors: | Wen, T.L, Du, M, Yang, X, Shen, Y.Q. | | Deposit date: | 2024-06-19 | | Release date: | 2025-03-05 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis of beta-arrestin coupling to the metabotropic glutamate receptor mGlu3.
Nat.Chem.Biol., 21, 2025
|
|
9IKJ
 
 | | Cryo-EM structure of TLP-1a | | Descriptor: | TLP-1 | | Authors: | Yan, N, Yan, C, Li, Z, Wang, T. | | Deposit date: | 2024-06-27 | | Release date: | 2024-11-13 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | CryoSeek: A strategy for bioentity discovery using cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9IKK
 
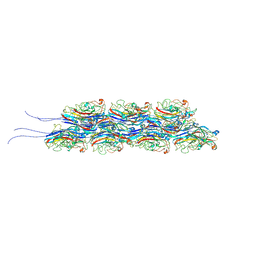 | | Cryo-EM structure of TLP-1b | | Descriptor: | TLP-2 | | Authors: | Yan, N, Yan, C, Li, Z, Wang, T. | | Deposit date: | 2024-06-27 | | Release date: | 2024-11-13 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | CryoSeek: A strategy for bioentity discovery using cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9IKM
 
 | | Cryo-EM structure of TLP-4 | | Descriptor: | TLP-4, alpha-L-arabinofuranose-(1-2)-alpha-L-arabinofuranose-(1-3)-alpha-L-arabinofuranose-(1-2)-alpha-D-mannopyranose-(1-2)-[beta-L-arabinofuranose-(1-2)-alpha-L-arabinofuranose-(1-3)-alpha-L-arabinofuranose-(1-3)]alpha-L-arabinofuranose-(1-4)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-L-arabinofuranose-(1-2)]beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-2)]beta-D-mannopyranose-(1-4)-[alpha-L-arabinofuranose-(1-2)]beta-D-galactopyranose, alpha-L-arabinofuranose-(1-2)-beta-D-galactopyranose-(1-2)-beta-L-arabinofuranose-(1-2)-[alpha-L-arabinofuranose-(1-2)-beta-L-arabinofuranose-(1-3)]beta-L-arabinofuranose-(1-3)-alpha-L-arabinofuranose, ... | | Authors: | Yan, N, Yan, C, Li, Z, Wang, T. | | Deposit date: | 2024-06-27 | | Release date: | 2025-01-08 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoSeek II: Cryo-EM analysis of glycofibrils from freshwater reveals well-structured glycans coating linear tetrapeptide repeats.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ISG
 
 | | Structure of rat TRPV1 in complex with PSFL426-S5 | | Descriptor: | 4-azanyl-3-sulfanylidene-1,2,4-triazine-5,6-dione, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Chen, X, Yu, Y. | | Deposit date: | 2024-07-17 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of capsaicin entry into buried vanilloid sites in TRPV1.
Nat.Chem.Biol., 2025
|
|
9IVY
 
 | |
9IXY
 
 | | Human KCNQ2-CaM-Ebio2 Complex in the Presence of PIP2 | | Descriptor: | Calmodulin-3, Isoform 3 of Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Yang, Z, Guo, J. | | Deposit date: | 2024-07-29 | | Release date: | 2025-03-19 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Small molecule inhibits KCNQ channels with a non-blocking mechanism.
Nat.Chem.Biol., 21, 2025
|
|
