8SIR
 
 | |
8SIS
 
 | |
8SIT
 
 | |
8SKQ
 
 | | RNA oligonucleotide containing an alpha-(L)-threofuranosyl nucleic acid (TNA) | | Descriptor: | TNA-containing RNA oligonucleotide | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Shorter Is Better: The alpha-(l)-Threofuranosyl Nucleic Acid Modification Improves Stability, Potency, Safety, and Ago2 Binding and Mitigates Off-Target Effects of Small Interfering RNAs.
J.Am.Chem.Soc., 145, 2023
|
|
8T1U
 
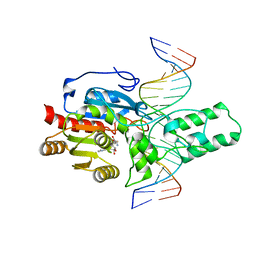 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
8T9C
 
 | |
8T9E
 
 | |
8T9X
 
 | |
8TA7
 
 | |
8TJE
 
 | |
8UBS
 
 | | Crystal structure of NrdJ-1 split intein fusion | | Descriptor: | FORMIC ACID, NrdJ-1, SODIUM ION | | Authors: | Kofoed, C, Ye, X, Jeffrey, P.D, Muir, T.W. | | Deposit date: | 2023-09-24 | | Release date: | 2024-10-09 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Programmable protein ligation on cell surfaces
Nature, 2025
|
|
8UUI
 
 | | X-ray structure of Interleukin-23 in complex with peptide 23-446 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joseph, J.S, Sridhar, V.S, Liu, M, Hu, Y, Gatchalian, J. | | Deposit date: | 2023-11-01 | | Release date: | 2025-07-09 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of an Induced Orthosteric Pocket in IL-23: A New Avenue for Non-biological Therapeutic Targeting.
Acs Chem.Biol., 20, 2025
|
|
8VAW
 
 | |
8VAX
 
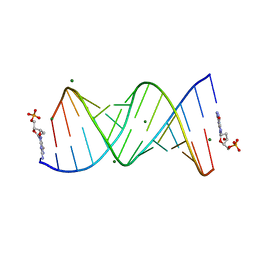 | |
8VK6
 
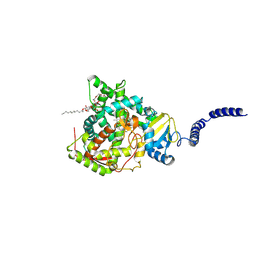 | | 14alpha-demethylase (CYP51) with amide-linked long arm extension antifungal azole inhibitor | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Lanosterol 14-alpha demethylase, N-[(2S)-2-(2,4-difluorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]-4'-(trifluoromethoxy)[1,1'-biphenyl]-4-carboxamide, ... | | Authors: | Tyndall, J.D.A, Monk, B.C, Simons, C. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Exploring Long Arm Amide-Linked Side Chains in the Design of Antifungal Azole Inhibitors of Sterol 14 alpha-Demethylase (CYP51).
J.Med.Chem., 68, 2025
|
|
8VWE
 
 | | HIV-1 neutralizing antibody D5_AR bound to HIV-1 gp41 coiled-coil pocket IQN17 | | Descriptor: | Neutralizing antibody D5_AR Heavy Chain, Neutralizing antibody D5_AR Light Chain, Transmembrane protein gp41 IQN17 peptide | | Authors: | Filsinger Interrante, M.V, Tang, S, Fernandez, D, Kim, P.S. | | Deposit date: | 2024-02-01 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Utilizing Machine Learning to Improve Neutralization Potency of an HIV-1 Antibody Targeting the gp41 N-Heptad Repeat.
Acs Chem.Biol., 20, 2025
|
|
8VYA
 
 | | SARS-CoV-2 Omicron Variant Spike Glycoprotein Fusion Core (Q954H) | | Descriptor: | SARS-CoV-2 Omicron variant spike glycoprotein C-terminal heptad repeat domain, SARS-CoV-2 Omicron variant spike glycoprotein N-terminal heptad repeat domain (Q954H) | | Authors: | Outlaw, V.K, Apurba, R, Vithanage, N. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-19 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | SARS-CoV-2 Omicron Variant Spike Glycoprotein Mutation Q954H Enhances Fusion Core Stability.
Acs Chem.Biol., 20, 2025
|
|
8WYH
 
 | |
8WYJ
 
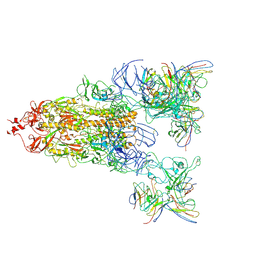 | | The global map of Omicron Subvariants Spike with two antibodies | | Descriptor: | S309-HC, S309-KC, SA55-HC, ... | | Authors: | Yan, R.H, Wang, A.J, Yang, H.N. | | Deposit date: | 2023-10-31 | | Release date: | 2024-11-13 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants.
Nat Commun, 15, 2024
|
|
8XMK
 
 | | Local refinement of SRCR5-9 domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2023-12-27 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Calcium-dependent oligomerization of scavenger receptor CD163 facilitates the endocytosis of ligands
Nat Commun, 16, 2025
|
|
8XMP
 
 | | Structure of CD163 in complex with Hb-Hp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2023-12-27 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Calcium-dependent oligomerization of scavenger receptor CD163 facilitates the endocytosis of ligands
Nat Commun, 16, 2025
|
|
8XMQ
 
 | | Structure of dimeric CD163 in complex with Hb-Hp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2023-12-27 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Calcium-dependent oligomerization of scavenger receptor CD163 facilitates the endocytosis of ligands
Nat Commun, 16, 2025
|
|
8XMW
 
 | | Local refinement of Hb-Hp bind to CD163 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2023-12-28 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Calcium-dependent oligomerization of scavenger receptor CD163 facilitates the endocytosis of ligands
Nat Commun, 16, 2025
|
|
8XP6
 
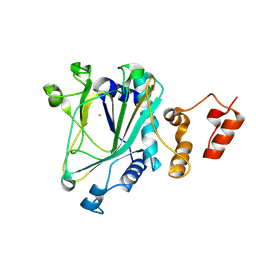 | | 2OG-Fe(II) oxygenase-ColD | | Descriptor: | 2OG-Fe(II) oxygenase, FE (III) ION | | Authors: | Zhang, B, Shi, J, Zhang, Y, Ge, H.M. | | Deposit date: | 2024-01-03 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biosynthesis of Collinodin Unveils Iterative Oxidative and Prenylation Modifications
Acs Catalysis, 15, 2025
|
|
8XPF
 
 | | 2OG-Fe(II) oxygenase-ColD in complex with collinodins | | Descriptor: | 2-OXOGLUTARIC ACID, 2OG-Fe(II) oxygenase, FE (III) ION, ... | | Authors: | Zhang, B, Shi, J, Zhang, Y, Ge, H.M. | | Deposit date: | 2024-01-03 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of Collinodin Unveils Iterative Oxidative and Prenylation Modifications
Acs Catalysis, 15, 2025
|
|
