8ID0
 
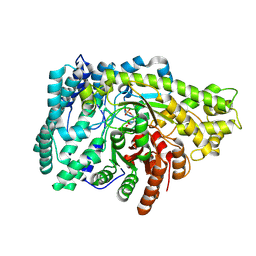 | | Crystal structure of PflD bound to 1,5-anhydromannitol-6-phosphate in Streptococcus dysgalactiae subsp. equisimilis | | Descriptor: | [(2R,3S,4R,5R)-3,4,5-tris(oxidanyl)oxan-2-yl]methyl dihydrogen phosphate, formate C-acetyltransferase | | Authors: | Ma, K.L, Zhang, Y. | | Deposit date: | 2023-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 2024
|
|
8ID7
 
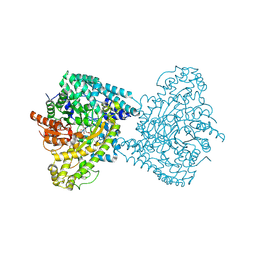 | |
8OH6
 
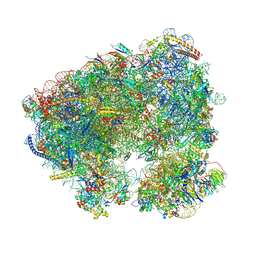 | | Crystal structure of the Candida albicans 80S ribosome in complex with Paromomycin (500umol) | | Descriptor: | 18S, 25S (gene name XR_002086444.1), 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
8OS1
 
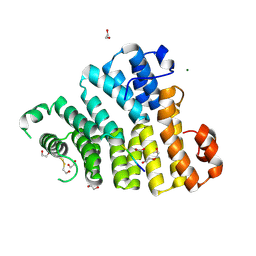 | | X-ray structure of the Peroxisomal Targeting Signal 1 (PTS1) of Trypanosoma Cruzi PEX5 in complex with the PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Napolitano, V, Blat, A, Popowicz, G.M, Dubin, G. | | Deposit date: | 2023-04-17 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dynamics of the TPR domain of the peroxisomal cargo receptor Pex5 in Trypanosoma.
Int.J.Biol.Macromol., 2024
|
|
8PWC
 
 | | Crystal structure of MDM2 with Brigimadlin | | Descriptor: | Brigimadlin, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Bader, G, Wolkerstorfer, B. | | Deposit date: | 2023-07-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Discovery and Characterization of Brigimadlin, a Novel and Highly Potent MDM2-p53 Antagonist Suitable for Intermittent Dose Schedules.
Mol.Cancer Ther., 2024
|
|
8QKW
 
 | | Crystal structure of Levansucrase from Pseudomonas syringae in complex with a tetravalent iminosugar | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-(hydroxymethyl)-1-[6-[4-[[3-[[3-[6-[(2~{S},3~{R},4~{S})-2-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-1-yl]hexyl]-1,2,3-triazol-4-yl]methoxy]-2,2-bis[[1-[6-[2-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-1-yl]hexyl]-1,2,3-triazol-4-yl]methoxymethyl]propoxy]methyl]-1,2,3-triazol-1-yl]hexyl]pyrrolidine-3,4-diol, DIMETHYL SULFOXIDE, ... | | Authors: | Ferraroni, M, Canovai, A. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Levansucrase from Pseudomonas syringae in complex with a tetravalent iminosugar
To Be Published
|
|
8QKZ
 
 | | Human Carbonic Anhydrase II in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol at pH 9.0 | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, 1,2-ETHANEDIOL, Carbonic anhydrase 2, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Human Carbonic Anhydrase II in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol at pH 9.0
To Be Published
|
|
8QL3
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 233 fs structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL4
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 349 fs structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL5
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 1 ps structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL6
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 25 ps structure | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL7
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 35 ps structure | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL8
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 125 ps structure | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QLB
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 100 ns structure | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QLN
 
 | |
8QLU
 
 | |
8QLW
 
 | |
8QLX
 
 | |
8QMB
 
 | | Nucleant-assisted 2.0 A resolution structure of the Streptococcus pneumoniae topoisomerase IV-V18mer DNA complex with the novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2023-09-21 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
8QMC
 
 | | High resolution structure of the Streptococcus pneumoniae topoisomerase IV-complex with the V-site 18mer dsDNA and novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2023-09-21 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
8QMJ
 
 | |
8QMQ
 
 | |
8QMR
 
 | |
8QMS
 
 | |
8QMT
 
 | |
