8VZF
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase ApbHACS from Alphaproteobacteria bacterium in the Complex with THDP, Coenzyme A, and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-Hydroxyacyl-CoA Lyase/Synthase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gade, P, Kim, Y, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZH
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA lyase/synthase TbHACS from Thermoflexaceae bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, M, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZI
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase CcHACS from Conidiobolus coronatus in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxyacyl-CoA lyase-like protein 1, ACETATE ION, ... | | Authors: | Kim, Y, Gade, P, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZJ
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase DhcHACS from Dehalococcoidia bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Kim, Y, Gade, P, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZK
 
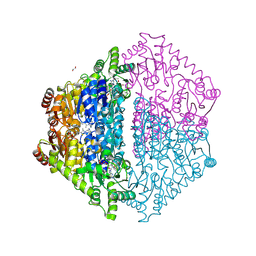 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase CfhHACS from Chloroflexi bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Gade, P, Mayfield, A, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8W7R
 
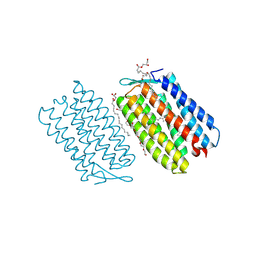 | | H. walsbyi bacteriorhodopsin mutant - W94F | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin-I, GLYCEROL, ... | | Authors: | Li, G.Y, Chen, J.C, Yang, C.S. | | Deposit date: | 2023-08-31 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of H. walsbyi bacteriorhodopsin mutant - W94F at 2.38 Angstroms resolution.
To Be Published
|
|
8WIP
 
 | |
8WJ5
 
 | | Double-wing domain of E. coli YdcD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Uncharacterized protein YdcD | | Authors: | Kamata, K, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Making the unphasable feasible
To Be Published
|
|
8WJB
 
 | |
8WJW
 
 | |
8WJZ
 
 | | Crystal structure of AtHPPD-F419Y+Y13161 complex | | Descriptor: | 3-(2,6-dimethylphenyl)-1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Crystal structure of AtHPPD-F419Y+Y13161 complex
To Be Published
|
|
8WK1
 
 | |
8WK2
 
 | | Crystal structure of AtHPPD-F419Y+Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Crystal structure of AtHPPD-F419Y+Y13287 complex
To Be Published
|
|
8WK5
 
 | |
8WK7
 
 | |
8WK9
 
 | |
8WKA
 
 | |
8WKB
 
 | |
8WKC
 
 | | Crystal structure of OgBVMO(Oceanicola granulosus) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Du, Y, Wang, Y.H. | | Deposit date: | 2023-09-27 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of OgBVMO(Oceanicola granulosus)
To be published
|
|
8WKE
 
 | |
8WKF
 
 | |
8WKG
 
 | |
8WL0
 
 | | Crystal structure of AtHPPD+Y19542 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-3-[(4-fluorophenyl)methyl]-5-methyl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-29 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of AtHPPD-+Y19542 complex
To Be Published
|
|
8WL1
 
 | |
8WL3
 
 | |
