1A8V
 
 | |
1A8Z
 
 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1A95
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH MG:CPRPP AND GUANINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, GUANINE, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1A96
 
 | | XPRTASE FROM E. COLI WITH BOUND CPRPP AND XANTHINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1A97
 
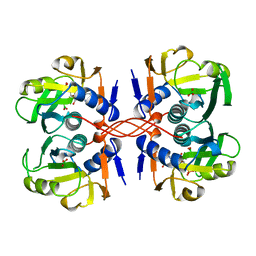 | | XPRTASE FROM E. COLI COMPLEXED WITH GMP | | Descriptor: | BORIC ACID, GUANOSINE-5'-MONOPHOSPHATE, XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1A98
 
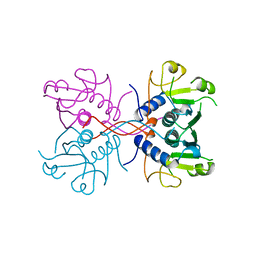 | | XPRTASE FROM E. COLI COMPLEXED WITH GMP | | Descriptor: | XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1A99
 
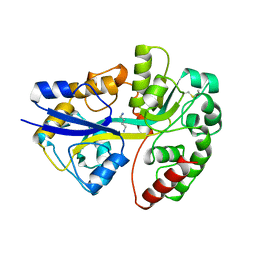 | | PUTRESCINE RECEPTOR (POTF) FROM E. COLI | | Descriptor: | 1,4-DIAMINOBUTANE, PUTRESCINE-BINDING PROTEIN | | Authors: | Vassylyev, D.G, Tomitori, H, Kashiwagi, K, Morikawa, K, Igarashi, K. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-21 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and mutational analysis of the Escherichia coli putrescine receptor. Structural basis for substrate specificity.
J.Biol.Chem., 273, 1998
|
|
1A9B
 
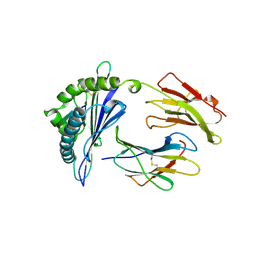 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-03 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
1A9E
 
 | | DECAMER-LIKE CONFORMATION OF A NANO-PEPTIDE BOUND TO HLA-B3501 DUE TO NONSTANDARD POSITIONING OF THE C-TERMINUS | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B*3501 (ALPHA CHAIN), ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1998-04-05 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus.
J.Mol.Biol., 285, 1999
|
|
1A9N
 
 | |
1A9W
 
 | | HUMAN EMBRYONIC GOWER II CARBONMONOXY HEMOGLOBIN | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), ... | | Authors: | Sutherland-Smith, A.J, Baker, H.M, Hofmann, O.M, Brittain, T, Baker, E.N. | | Deposit date: | 1998-04-11 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a human embryonic haemoglobin: the carbonmonoxy form of gower II (alpha2 epsilon2) haemoglobin at 2.9 A resolution.
J.Mol.Biol., 280, 1998
|
|
1AA0
 
 | | FIBRITIN DELETION MUTANT E (BACTERIOPHAGE T4) | | Descriptor: | CHLORIDE ION, FIBRITIN, ZINC ION | | Authors: | Tao, Y, Strelkov, S.V, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 1997-01-18 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain.
Structure, 5, 1997
|
|
1AA1
 
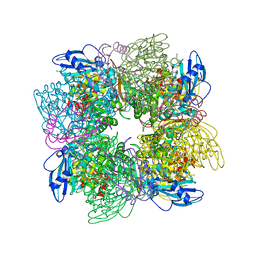 | |
1AA2
 
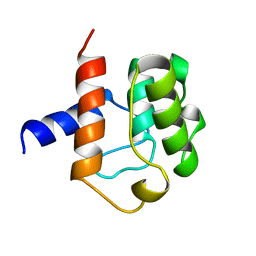 | |
1AA4
 
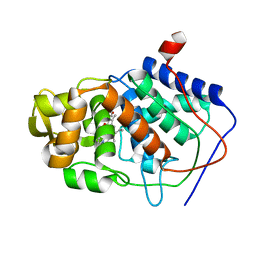 | | SPECIFICITY OF LIGAND BINDING IN A BURIED POLAR CAVITY OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1AA5
 
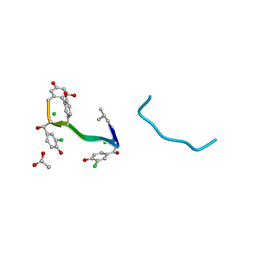 | | VANCOMYCIN | | Descriptor: | ACETIC ACID, CHLORIDE ION, VANCOMYCIN, ... | | Authors: | Loll, P.J, Bevivino, A.E, Korty, B.D, Axelsen, P.H. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Simultaneous Recognition of a Carboxylate-Containing Ligand and an Intramolecular Surrogate Ligand in the Crystal Structure of an Asymmetric Vancomycin Dimer.
J.Am.Chem.Soc., 119, 1997
|
|
1AA6
 
 | | REDUCED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
1AA7
 
 | |
1AAC
 
 | | AMICYANIN OXIDIZED, 1.31 ANGSTROMS | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1995-09-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | X-ray structure of the cupredoxin amicyanin, from Paracoccus denitrificans, refined at 1.31 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1AAL
 
 | |
1AAR
 
 | | STRUCTURE OF A DIUBIQUITIN CONJUGATE AND A MODEL FOR INTERACTION WITH UBIQUITIN CONJUGATING ENZYME (E2) | | Descriptor: | DI-UBIQUITIN | | Authors: | Cook, W.J, Jeffrey, L.C, Carson, M, Chen, Z, Pickart, C.M. | | Deposit date: | 1992-04-17 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2).
J.Biol.Chem., 267, 1992
|
|
1AAZ
 
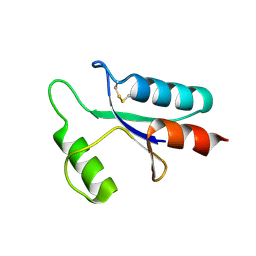 | | THE STRUCTURE OF OXIDIZED BACTERIOPHAGE T4 GLUTAREDOXIN (THIOREDOXIN) | | Descriptor: | CADMIUM ION, GLUTAREDOXIN | | Authors: | Eklund, H, Ingelman, M, Soderberg, B.-O, Uhlin, T, Nordlund, P, Nikkola, M, Sonnerstam, U, Joelson, T, Petratos, K. | | Deposit date: | 1992-04-24 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of oxidized bacteriophage T4 glutaredoxin (thioredoxin). Refinement of native and mutant proteins.
J.Mol.Biol., 228, 1992
|
|
1AB0
 
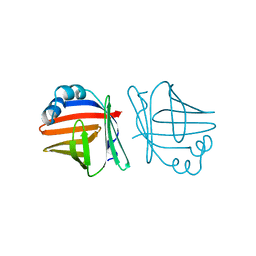 | | C1G/V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN AT PH 4.5 | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C, Simpson, M, Banaszak, L, Bernlohr, D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
1AB1
 
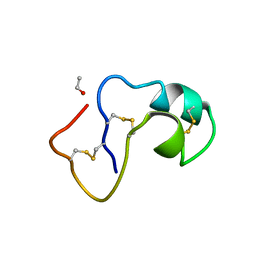 | | SI FORM CRAMBIN | | Descriptor: | CRAMBIN (SER22/ILE25), ETHANOL | | Authors: | Teeter, M.M, Yamano, A. | | Deposit date: | 1997-01-31 | | Release date: | 1997-08-12 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal structure of Ser-22/Ile-25 form crambin confirms solvent, side chain substate correlations.
J.Biol.Chem., 272, 1997
|
|
1AB4
 
 | |
