1SFR
 
 | | Crystal Structure of the Mycobacterium tuberculosis Antigen 85A Protein | | Descriptor: | Antigen 85-A | | Authors: | Ronning, D.R, Vissa, V, Besra, G.S, Belisle, J.T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-20 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis Antigen 85A and 85C Structures Confirm Binding Orientation and Conserved Substrate Specificity
J.Biol.Chem., 279, 2004
|
|
1SFS
 
 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
1SFU
 
 | | Crystal structure of the viral Zalpha domain bound to left-handed Z-DNA | | Descriptor: | 34L protein, 5'-D(*T*CP*GP*CP*GP*CP*G)-3' | | Authors: | Ha, S.C, Van Quyen, D, Wu, C.A, Lowenhaupt, K, Rich, A, Kim, Y.G, Kim, K.K. | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SFX
 
 | | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Conserved hypothetical protein AF2008 | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus
To be Published
|
|
1SFY
 
 | | Crystal structure of recombinant Erythrina corallodandron Lectin | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Kulkarni, K.A, Srivastava, A, Mitra, N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2004-02-21 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effect of glycosylation on the structure of Erythrina corallodendron lectin.
Proteins, 56, 2004
|
|
1SG0
 
 | | Crystal structure analysis of QR2 in complex with resveratrol | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NRH dehydrogenase [quinone] 2, RESVERATROL, ... | | Authors: | Buryanovskyy, L, Fu, Y, Boyd, M, Ma, Y, Tsieh, T.C, Wu, J.M, Zhang, Z. | | Deposit date: | 2004-02-22 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of quinone reductase 2 in complex with resveratrol
Biochemistry, 43, 2004
|
|
1SG1
 
 | |
1SG2
 
 | |
1SG3
 
 | | Structure of allantoicase | | Descriptor: | Allantoicase | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, Graille, M, Meyer, P, Liger, D, Blondeau, K, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2004-02-23 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Yeast Allantoicase Reveals a Repeated Jelly Roll Motif
J.Biol.Chem., 279, 2004
|
|
1SG4
 
 | | Crystal structure of human mitochondrial delta3-delta2-enoyl-CoA isomerase | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, mitochondrial, OCTANOYL-COENZYME A | | Authors: | Partanen, S.T, Novikov, D.K, Popov, A.N, Mursula, A.M, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2004-02-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of human mitochondrial Delta3-Delta2-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group.
J.Mol.Biol., 342, 2004
|
|
1SG6
 
 | | Crystal structure of Aspergillus nidulans 3-dehydroquinate synthase (AnDHQS) in complex with Zn2+ and NAD+, at 1.7D | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentafunctional AROM polypeptide, ZINC ION | | Authors: | Nichols, C.E, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the 'open' form of Aspergillus nidulans 3-dehydroquinate synthase at 1.7 A resolution from crystals grown following enzyme turnover.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SG8
 
 | | Crystal structure of the procoagulant fast form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1SG9
 
 | | Crystal structure of Thermotoga maritima protein HEMK, an N5-glutamine methyltransferase | | Descriptor: | GLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel mode of dimerization via formation of a glutamate anhydride crosslink in a protein crystal structure.
Proteins, 71, 2008
|
|
1SGF
 
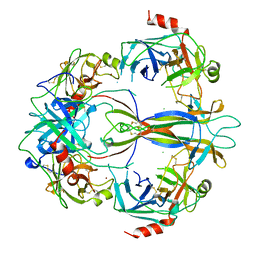 | | CRYSTAL STRUCTURE OF 7S NGF: A COMPLEX OF NERVE GROWTH FACTOR WITH FOUR BINDING PROTEINS (SERINE PROTEINASES) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NERVE GROWTH FACTOR, ... | | Authors: | Bax, B.D.V, Blundell, T.L, Murray-Rust, J, Mcdonald, N.Q. | | Deposit date: | 1997-08-08 | | Release date: | 1998-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of mouse 7S NGF: a complex of nerve growth factor with four binding proteins.
Structure, 5, 1997
|
|
1SGH
 
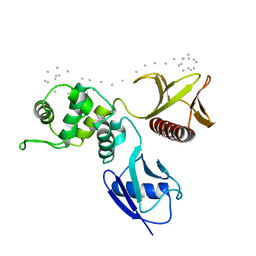 | | Moesin FERM domain bound to EBP50 C-terminal peptide | | Descriptor: | Ezrin-radixin-moesin binding phosphoprotein 50, Moesin | | Authors: | Finnerty, C.M, Chambers, D, Ingraffea, J, Faber, H.R, Karplus, P.A, Bretscher, A. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain
J.Cell.Sci., 117, 2004
|
|
1SGI
 
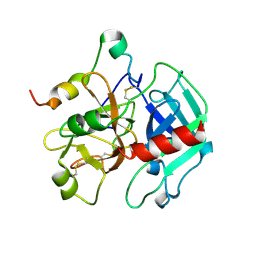 | | Crystal structure of the anticoagulant slow form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1SGK
 
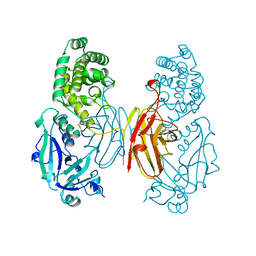 | | NUCLEOTIDE-FREE DIPHTHERIA TOXIN | | Descriptor: | DIPHTHERIA TOXIN (DIMERIC) | | Authors: | Bell, C.E, Eisenberg, D. | | Deposit date: | 1996-09-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of nucleotide-free diphtheria toxin.
Biochemistry, 36, 1997
|
|
1SGL
 
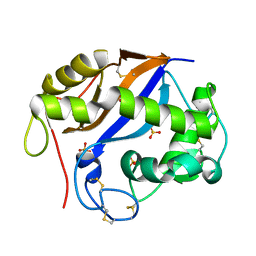 | | The three-dimensional structure and X-ray sequence reveal that trichomaglin is a novel S-like ribonuclease | | Descriptor: | SULFATE ION, trichomaglin | | Authors: | Gan, J.-H, Yu, L, Wu, J, Xu, H, Choudhary, J.S, Blackstock, W.P, Liu, W.-Y, Xia, Z.-X. | | Deposit date: | 2004-02-24 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure and X-ray sequence reveal that trichomaglin is a novel S-like ribonuclease.
Structure, 12, 2004
|
|
1SGM
 
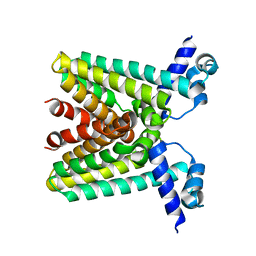 | |
1SGU
 
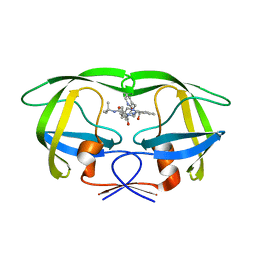 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, Mckenna, R, Abandje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
1SGV
 
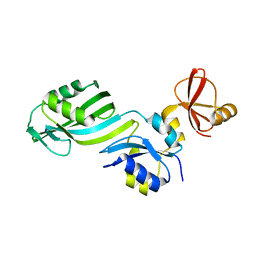 | | STRUCTURE OF TRNA PSI55 PSEUDOURIDINE SYNTHASE (TRUB) | | Descriptor: | tRNA pseudouridine synthase B | | Authors: | Chaudhuri, B.N, Chan, S, Perry, L.J, Yeates, T.O, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the apo forms of psi 55 tRNA pseudouridine synthase from Mycobacterium tuberculosis: a hinge at the base of the catalytic cleft.
J.Biol.Chem., 279, 2004
|
|
1SGW
 
 | | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001 | | Descriptor: | CHLORIDE ION, SODIUM ION, putative ABC transporter | | Authors: | Liu, Z.J, Tempel, W, Shah, A, Chen, L, Lee, D, Kelley, L.-L.C, Dillard, B.D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001
To be Published
|
|
1SH0
 
 | | Crystal Structure of Norwalk Virus Polymerase (Triclinic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1SH2
 
 | | Crystal Structure of Norwalk Virus Polymerase (Metal-free, Centered Orthorhombic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1SH3
 
 | | Crystal Structure of Norwalk Virus Polymerase (MgSO4 crystal form) | | Descriptor: | MAGNESIUM ION, RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
